4PRN
 
 | | Crystal structure of a HLA-B*35:01-HPVG-A4 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-06 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PRE
 
 | | Crystal structure of a HLA-B*35:08-HPVG-Q5 | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, MHC class I antigen | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PRB
 
 | | Crystal structure of a HLA-B*35:08-HPVG-A4 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PR5
 
 | | Crystal structure of a HLA-B*35:01-HPVG-D5 | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, HLA class I histocompatibility antigen, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PRI
 
 | | Crystal structure of TK3 TCR-HLA-B*35:08-HPVG complex | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, Epstein-Barr nuclear antigen 1, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PRP
 
 | | Crystal structure of TK3 TCR-HLA-B*35:01-HPVG-Q5 complex | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, MHC class I antigen, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-06 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PRA
 
 | | Crystal structure of a HLA-B*35:01-HPVG-Q5 | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, MHC class I antigen | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PRH
 
 | | Crystal structure of TK3 TCR-HLA-B*35:08-HPVG-D5 complex | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 1, MHC class I antigen, ... | | Authors: | Yu Chih, L, Rossjohn, J, Gras, S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Molecular Basis for the Interplay between T Cells, Viral Mutants, and Human Leukocyte Antigen Micropolymorphism.
J.Biol.Chem., 289, 2014
|
|
4PL6
 
 | |
3KXF
 
 | | Crystal Structure of SB27 TCR in complex with the 'restriction triad' mutant HLA-B*3508-13mer | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Archbold, J.K, Tynan, F.E, Gras, S, Rossjohn, J. | | Deposit date: | 2009-12-03 | | Release date: | 2010-06-09 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KWW
 
 | | Crystal structure of the 'restriction triad' mutant of HLA B*3508, beta-2-microglobulin and EBV peptide | | Descriptor: | ACETIC ACID, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Archbold, J.K, Tynan, F.E, Gras, S, Rossjohn, J. | | Deposit date: | 2009-12-01 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability
Proc.Natl.Acad.Sci.USA, 2010
|
|
5XRY
 
 | |
2PJF
 
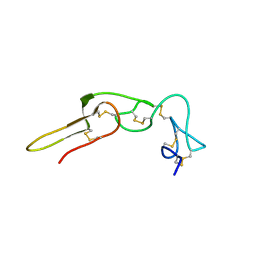 | | Solution structure of rhodostomin | | Descriptor: | Rhodostoxin-disintegrin rhodostomin | | Authors: | Chuang, W.J, Chen, Y.C, Chen, C.Y, Chang, Y.T. | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Effect of D to E mutation of the RGD motif in rhodostomin on its activity, structure, and dynamics: Importance of the interactions between the D residue and integrin
Proteins, 2009
|
|
2PJG
 
 | | Solution structure of rhodostomin D51E mutant | | Descriptor: | Rhodostoxin-disintegrin rhodostomin | | Authors: | Chuang, W.J, Chen, Y.C, Chen, C.Y, Chou, L.J. | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-08 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Effect of D to E mutation of the RGD motif in rhodostomin on its activity, structure, and dynamics: Importance of the interactions between the D residue and integrin
Proteins, 2009
|
|
6VHI
 
 | |
8E19
 
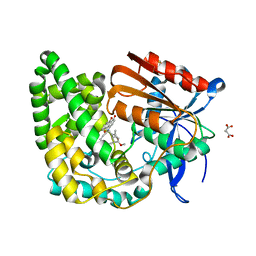 | | Crystal structure of TnmK1 complexed with TNM H | | Descriptor: | (1R,8S,13S)-8-[(4-hydroxy-9,10-dioxo-9,10-dihydroanthracen-1-yl)amino]-12-methoxy-10-methylbicyclo[7.3.1]trideca-9,11-diene-2,6-diyne-13-carbaldehyde, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SUCCINIC ACID, ... | | Authors: | Liu, Y.-C, Gui, C, Shen, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Intramolecular C-C Bond Formation Links Anthraquinone and Enediyne Scaffolds in Tiancimycin Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
8E18
 
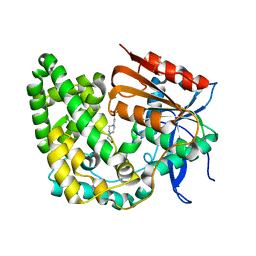 | | Crystal structure of apo TnmK1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Secreted hydrolase | | Authors: | Liu, Y.-C, Gui, C, Shen, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Intramolecular C-C Bond Formation Links Anthraquinone and Enediyne Scaffolds in Tiancimycin Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
4YHA
 
 | |
4YGF
 
 | | Crystal structure of the complex of Helicobacter pylori alpha-Carbonic Anhydrase with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Alpha-carbonic anhydrase, CHLORIDE ION, ... | | Authors: | Roujeinikova, A, Modak, J.K. | | Deposit date: | 2015-02-26 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Inhibition of Helicobacter pylori alpha-Carbonic Anhydrase by Sulfonamides.
Plos One, 10, 2015
|
|
5GI5
 
 | |
5GI7
 
 | | Crystal Structure of Drosophila melanogaster Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-phenylethylamine | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, COENZYME A, Dopamine N-acetyltransferase, ... | | Authors: | Yang, Y.C, Lin, S.J, Cheng, K.C, Cheng, H.C, Lyu, P.C. | | Deposit date: | 2016-06-22 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | An essential role of acetyl coenzyme A in the catalytic cycle of insect arylalkylamine N-acetyltransferase.
Commun Biol, 3, 2020
|
|
5GI9
 
 | | Crystal Structure of Drosophila melanogaster Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-Tryptamine | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, COENZYME A, Dopamine N-acetyltransferase, ... | | Authors: | Yang, Y.C, Lin, S.J, Cheng, K.C, Cheng, H.C, Lyu, P.C. | | Deposit date: | 2016-06-22 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An essential role of acetyl coenzyme A in the catalytic cycle of insect arylalkylamine N-acetyltransferase.
Commun Biol, 3, 2020
|
|
5GUT
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - N1248A | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Chen, S.J, Ye, F. | | Deposit date: | 2016-08-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
5GUV
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - R1279D | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, ZINC ION | | Authors: | Ye, F, Chen, S.J. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.078 Å) | | Cite: | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
4QR5
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | Bromodomain-containing protein 4, N-[3-(cyclopentylsulfamoyl)-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]cyclopropanecarboxamide | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
