8BEY
 
 | | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein at pH 7 | | Descriptor: | Cry49Aa protein | | Authors: | Williamson, L.J, Rizkallah, P.J, Berry, C, Oberthur, D, Galchenkova, M, Yefanov, O, Bean, R. | | Deposit date: | 2022-10-22 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated from natural crystals using MHz-SFX.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BEZ
 
 | | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein at pH 11 | | Descriptor: | Cry49Aa protein | | Authors: | Williamson, L.J, Rizkallah, P.J, Berry, C, Oberthur, D, Galchenkova, M, Yefanov, O, Bean, R. | | Deposit date: | 2022-10-22 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Lysinibacillus sphaericus Tpp49Aa1 pesticidal protein elucidated from natural crystals using MHz-SFX.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6R5V
 
 | |
6R5P
 
 | |
6R5U
 
 | |
6R5T
 
 | |
6R5N
 
 | |
6R5O
 
 | |
6R5I
 
 | |
6R5R
 
 | |
7BYO
 
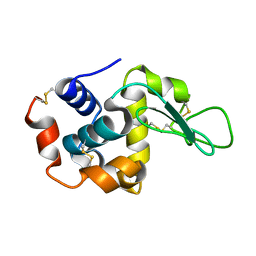 | | Lysozyme structure SS1 from SS mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
7BYP
 
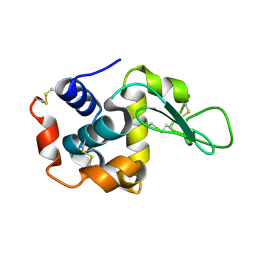 | | Lysozyme structure SASE1 from SASE mode | | Descriptor: | Lysozyme C | | Authors: | Kang, H.S, Lee, S.J. | | Deposit date: | 2020-04-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-brightness self-seeded X-ray free-electron laser covering the 3.5 keV to 14.6 keV range
Nat Photonics, 2021
|
|
8BRE
 
 | |
8CPB
 
 | | 1,6-anhydro-n-actetylmuramic acid kinase (AnmK) in complex with AMPPNP, and AnhMurNAc at 1.7 Angstroms resolution. | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, Anhydro-N-acetylmuramic acid kinase, GLYCEROL, ... | | Authors: | Jimenez-Faraco, E, Hermoso, J.A. | | Deposit date: | 2023-03-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic process of anhydro-N-acetylmuramic acid kinase from Pseudomonas aeruginosa.
J.Biol.Chem., 299, 2023
|
|
7O4B
 
 | |
7OK9
 
 | |
5X1E
 
 | |
5X1U
 
 | |
4Y4V
 
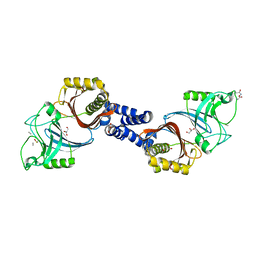 | | Structure of Helicobacter pylori Csd6 in the D-Ala-bound state | | Descriptor: | Conserved hypothetical secreted protein, D-ALANINE, GLYCEROL | | Authors: | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | Deposit date: | 2015-02-11 | | Release date: | 2015-09-02 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
5X1H
 
 | | Structure of Legionella pneumophila DotN | | Descriptor: | IcmJ (DotN), ZINC ION | | Authors: | Kwak, M.J, Oh, B.H. | | Deposit date: | 2017-01-26 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Architecture of the type IV coupling protein complex of Legionella pneumophila
Nat Microbiol, 2, 2017
|
|
5DYL
 
 | | Crystal structure of the cGMP-dependent protein kinase PKG from Plasmodium Vivax - Apo form | | Descriptor: | cGMP-dependent protein kinase, putative | | Authors: | Wernimont, A.K, Tempel, W, He, H, Seitova, A, Hills, T, Neculai, A.M, Baker, D.A, Flueck, C, Kettleborough, C.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-24 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5DYK
 
 | | Crystal structure of the cGMP-dependent protein kinase PKG from Plasmodium falciparum - Apo form | | Descriptor: | 1,2-ETHANEDIOL, CGMP-dependent protein kinase, GLYCEROL, ... | | Authors: | Wernimont, A.K, Tempel, W, He, H, Seitova, A, Hills, T, Neculai, A.M, Baker, D.A, Flueck, C, Kettleborough, C.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-24 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5DZC
 
 | | Crystal structure of the cGMP-dependent protein kinase PKG from Plasmodium Vivax - AMPPNP bound | | Descriptor: | CHLORIDE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SODIUM ION, ... | | Authors: | Walker, J.R, El Bakkouri, M, Loppnau, P, Graslund, S, He, H, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-25 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5E16
 
 | | Co-crystal structure of the N-termial cGMP binding domain of Plasmodium falciparum PKG with cGMP | | Descriptor: | CGMP-dependent protein kinase, CYCLIC GUANOSINE MONOPHOSPHATE | | Authors: | El Bakkouri, M, Walker, J.R, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6LIX
 
 | | CRL Protein of Arabidopsis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromophore lyase CRL, chloroplastic | | Authors: | Wang, F.F, Guan, K.L, Sun, P.K, Xing, W.M. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | The Arabidopsis CRUMPLED LEAF protein, a homolog of the cyanobacterial bilin lyase, retains the bilin-binding pocket for a yet unknown function.
Plant J., 104, 2020
|
|
