9ASS
 
 | |
9ASX
 
 | |
9ATK
 
 | |
9ATU
 
 | |
2GOM
 
 | |
2GOX
 
 | | Crystal structure of Efb-C / C3d Complex | | Descriptor: | Complement C3, Fibrinogen-binding protein | | Authors: | Hammel, M, Geisbrecht, B.V. | | Deposit date: | 2006-04-14 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structural basis for complement inhibition by Staphylococcus aureus.
Nat.Immunol., 8, 2007
|
|
4DPW
 
 | | Crystal structure of Staphylococcus epidermidis D283A mevalonate diphosphate decarboxylase complexed with mevalonate diphosphate and ATPgS | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Barta, M.L, McWhorter, W.J, Geisbrecht, B.V. | | Deposit date: | 2012-02-14 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural basis for nucleotide binding and reaction catalysis in mevalonate diphosphate decarboxylase.
Biochemistry, 51, 2012
|
|
4DU7
 
 | |
4DPY
 
 | |
4DPU
 
 | |
4DPX
 
 | |
4DU8
 
 | |
4DPT
 
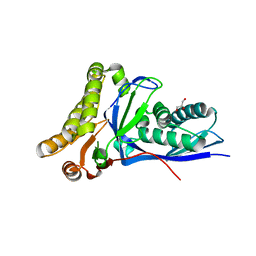 | | Crystal structure of Staphylococcus epidermidis mevalonate diphosphate decarboxylase complexed with inhibitor 6-FMVAPP and ATPgS | | Descriptor: | (3R)-3-(fluoromethyl)-3-hydroxy-5-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}pentanoic acid, GLYCEROL, Mevalonate diphosphate decarboxylase, ... | | Authors: | Barta, M.L, McWhorter, W.J, Geisbrecht, B.V. | | Deposit date: | 2012-02-14 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structural basis for nucleotide binding and reaction catalysis in mevalonate diphosphate decarboxylase.
Biochemistry, 51, 2012
|
|
3QT7
 
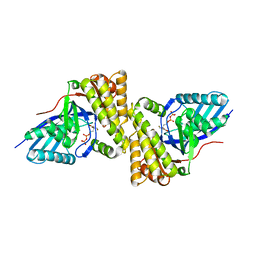 | | Crystal structure of Staphylococcus epidermidis mevalonate diphosphate decarboxylase complexed with inhibitor 6-FMVAPP | | Descriptor: | (3R)-3-(fluoromethyl)-3-hydroxy-5-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}pentanoic acid, Mevalonate diphosphate decarboxylase | | Authors: | Barta, M.L, Skaff, A.D, McWhorter, W.J, Miziorko, H.M, Geisbrecht, B.V. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Crystal structures of Staphylococcus epidermidis mevalonate diphosphate decarboxylase bound to inhibitory analogs reveal new insight into substrate binding and catalysis.
J.Biol.Chem., 286, 2011
|
|
3T46
 
 | |
3T49
 
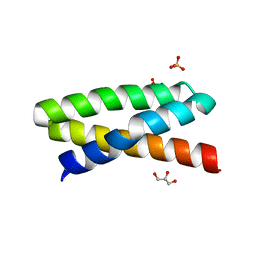 | |
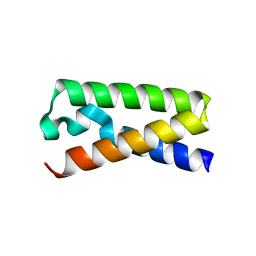
3T47
 
 | |
3QT6
 
 | | Crystal structure of Staphylococcus epidermidis mevalonate diphosphate decarboxylase complexed with inhibitor DPGP | | Descriptor: | 1-({[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}acetyl)-L-proline, Mevalonate diphosphate decarboxylase | | Authors: | Barta, M.L, Skaff, A.D, McWhorter, W.J, Miziorko, H.M, Geisbrecht, B.V. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Crystal structures of Staphylococcus epidermidis mevalonate diphosphate decarboxylase bound to inhibitory analogs reveal new insight into substrate binding and catalysis.
J.Biol.Chem., 286, 2011
|
|
3QT8
 
 | | Crystal structure of mutant S192A Staphylococcus epidermidis mevalonate diphosphate decarboxylase complexed with inhibitor 6-FMVAPP | | Descriptor: | (3R)-3-(fluoromethyl)-3-hydroxy-5-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}pentanoic acid, GLYCEROL, Mevalonate diphosphate decarboxylase | | Authors: | Barta, M.L, Skaff, A.D, McWhorter, W.J, Miziorko, H.M, Geisbrecht, B.V. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of Staphylococcus epidermidis mevalonate diphosphate decarboxylase bound to inhibitory analogs reveal new insight into substrate binding and catalysis.
J.Biol.Chem., 286, 2011
|
|
3T48
 
 | |
3R9V
 
 | | Cocrystal Structure of Proteolytically Truncated Form of IpaD from Shigella flexneri Bound to Deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCEROL, Invasin ipaD | | Authors: | Barta, M.L, Dickenson, N.E, Picking, W.L, Picking, W.D, Geisbrecht, B.V. | | Deposit date: | 2011-03-26 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of the bile salt binding site on IpaD from Shigella flexneri and the influence of ligand binding on IpaD structure.
Proteins, 80, 2012
|
|
3T4A
 
 | | Structure of a truncated form of Staphylococcal Complement Inhibitor B bound to human C3c at 3.4 Angstrom resolution | | Descriptor: | Complement C3 beta chain, Complement C3c alpha' chain fragment 1, Complement C3c alpha' chain fragment 2, ... | | Authors: | Garcia, B.L, Geisbrecht, B.V, Summers, B.J. | | Deposit date: | 2011-07-25 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Diversity in the C3b Convertase Contact Residues and Tertiary Structures of the Staphylococcal Complement Inhibitor (SCIN) Protein Family.
J.Biol.Chem., 287, 2012
|
|
3QT5
 
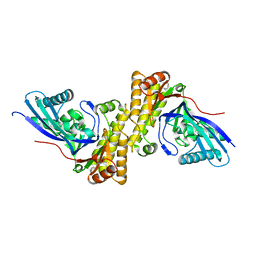 | | Crystal structure of Staphylococcus epidermidis mevalonate diphosphate decarboxylase | | Descriptor: | Mevalonate diphosphate decarboxylase | | Authors: | Barta, M.L, Skaff, A.D, McWhorter, W.J, Miziorko, H.M, Geisbrecht, B.V. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Crystal structures of Staphylococcus epidermidis mevalonate diphosphate decarboxylase bound to inhibitory analogs reveal new insight into substrate binding and catalysis.
J.Biol.Chem., 286, 2011
|
|
7RMU
 
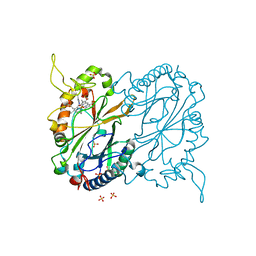 | |
5WKQ
 
 | | 2.10 A resolution structure of IpaB (residues 74-242) from Shigella flexneri | | Descriptor: | Invasin IpaB | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Picking, W.L, Picking, W.D. | | Deposit date: | 2017-07-25 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Using disruptive insertional mutagenesis to identify the in situ structure-function landscape of the Shigella translocator protein IpaB.
Protein Sci., 27, 2018
|
|
