4GYF
 
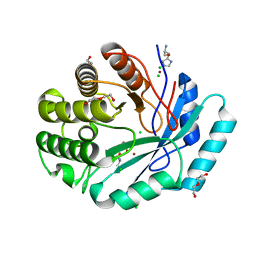 | | Crystal structure of histidinol phosphate phosphatase (HISK) from Lactococcus lactis subsp. lactis Il1403 complexed with ZN, L-histidinol and phosphate | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Histidinol-phosphatase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Ghodge, S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-09-05 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Crystal structure of histidinol phosphate phosphatase (HISK) from Lactococcus lactis subsp. lactis Il1403 complexed with ZN, L-histidinol and phosphate
To be Published
|
|
4HCD
 
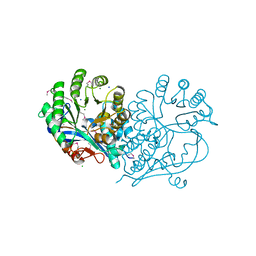 | | Crystal structure of D-glucarate dehydratase from agrobacterium tumefaciens complexed with magnesium | | Descriptor: | CHLORIDE ION, Isomerase/lactonizing enzyme, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Millikin, C, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-09-29 | | Release date: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of D-glucarate dehydratase from agrobacterium tumefaciens complexed with magnesium
To be Published
|
|
4HCL
 
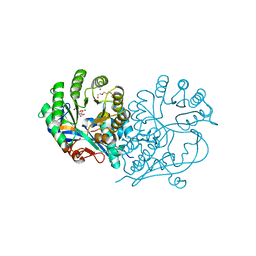 | | CRYSTAL STRUCTURE OF D-GLUCARATE DEHYDRATASE FROM AGROBACTERIUM TUMEFACIENS complexed with magnesium and L-Lyxarohydroxamate | | Descriptor: | (2R,3S,4R)-2,3,4-TRIHYDROXY-5-(HYDROXYAMINO)-5-OXOPENTANOIC ACID, Isomerase/lactonizing enzyme, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-09-30 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF D-GLUCARATE DEHYDRATASE FROM AGROBACTERIUM TUMEFACIENS complexed with magnesium and L-Lyxarohydroxamate
To be Published
|
|
4ICM
 
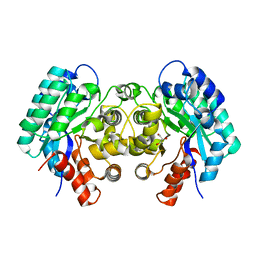 | | Crystal structure of 5-carboxyvanillate decarboxylase LigW from Sphingomonas paucimobilis | | Descriptor: | 5-carboxyvanillate decarboxylase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Vladimirova, A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-12-10 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.825 Å) | | Cite: | Crystal structure of 5-carboxyvanillate decarboxylase LigW from Sphingomonas paucimobilis
To be Published
|
|
4IP4
 
 | | Crystal structure of l-fuconate dehydratase from Silicibacter sp. tm1040 liganded with Mg | | Descriptor: | CARBON DIOXIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-01-09 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.128 Å) | | Cite: | Crystal structure of l-fuconate dehydratase from Silicibacter sp. tm1040 liganded with mg
To be Published
|
|
4IP5
 
 | | Crystal structure of l-fuconate dehydratase from Silicibacter sp. tm1040 liganded with Mg and d-erythronohydroxamate | | Descriptor: | (2R,3R)-N,2,3,4-TETRAHYDROXYBUTANAMIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-01-09 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of l-fuconate dehydratase from Silicibacter sp. tm1040 liganded with Mg and d-erythronohydroxamate
To be Published
|
|
1CFY
 
 | | YEAST COFILIN, MONOCLINIC CRYSTAL FORM | | Descriptor: | COFILIN | | Authors: | Fedorov, A.A, Lappalainen, P, Fedorov, E.V, Drubin, D.G, Almo, S.C. | | Deposit date: | 1996-11-26 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of yeast cofilin.
Nat.Struct.Biol., 4, 1997
|
|
1DFC
 
 | | CRYSTAL STRUCTURE OF HUMAN FASCIN, AN ACTIN-CROSSLINKING PROTEIN | | Descriptor: | FASCIN | | Authors: | Fedorov, A.A, Fedorov, E.V, Ono, S, Matsumura, F, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-11-18 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure, evolutionary conservation, and conformational dynamics of Homo sapiens fascin-1, an F-actin crosslinking protein.
J.Mol.Biol., 400, 2010
|
|
1COF
 
 | | YEAST COFILIN, ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | COFILIN | | Authors: | Fedorov, A.A, Lappalainen, P, Fedorov, E.V, Drubin, D.G, Almo, S.C. | | Deposit date: | 1996-11-26 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of yeast cofilin.
Nat.Struct.Biol., 4, 1997
|
|
1T62
 
 | |
1TZZ
 
 | | Crystal structure of the protein L1841, unknown member of enolase superfamily from Bradyrhizobium japonicum | | Descriptor: | Hypothetical protein L1841, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-12 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the protein L1841, unknown member of enolase superfamily from Bradyrhizobium japonicum
To be Published
|
|
1RC6
 
 | | Crystal structure of protein Ylba from E. coli, Pfam DUF861 | | Descriptor: | Hypothetical protein ylbA | | Authors: | Fedorov, A.A, Fedorov, E.V, Thirumuruhan, R, Ramagopal, U.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-03 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Ylba, hypothetical protein from E.Coli
To be Published
|
|
1SEF
 
 | |
1RVK
 
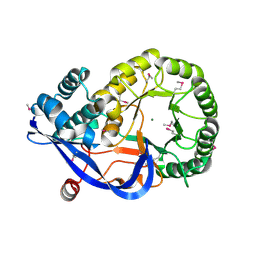 | | Crystal structure of enolase AGR_L_2751 from Agrobacterium Tumefaciens | | Descriptor: | MAGNESIUM ION, isomerase/lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Thirumuruhan, R, Zencheck, W, Millikin, C, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-12-14 | | Release date: | 2003-12-23 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of enzymatic activites in the Enolase superfamily: 1.7 A crystal structure of the hypothetical protein MR.GI-17937161 from Agrobacterium tumefaciens
To be Published
|
|
1TEL
 
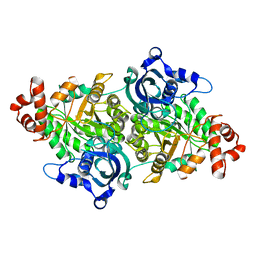 | | Crystal structure of a RubisCO-like protein from Chlorobium tepidum | | Descriptor: | ribulose bisphosphate carboxylase, large subunit | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-05-25 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a RubisCO-like protein from Chlorobium tepidum
To be Published
|
|
1SV6
 
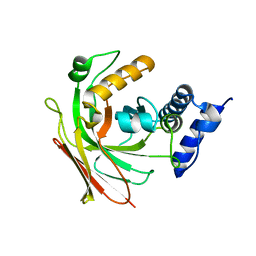 | | Crystal structure of 2-hydroxypentadienoic acid hydratase from Escherichia Coli | | Descriptor: | 2-keto-4-pentenoate hydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sharp, A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-03-28 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 2-hydroxypentadienoic acid hydratase from Escherichia Coli
To be Published
|
|
1SFN
 
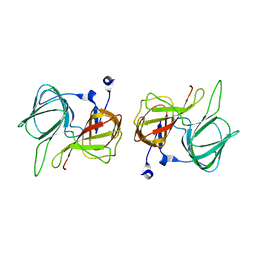 | |
2DW6
 
 | | Crystal structure of the mutant K184A of D-Tartrate Dehydratase from Bradyrhizobium japonicum complexed with Mg++ and D-tartrate | | Descriptor: | Bll6730 protein, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-08-07 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium japonicum
Biochemistry, 45, 2006
|
|
2DW7
 
 | | Crystal structure of D-tartrate dehydratase from Bradyrhizobium japonicum complexed with Mg++ and meso-tartrate | | Descriptor: | Bll6730 protein, MAGNESIUM ION, S,R MESO-TARTARIC ACID | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-08-07 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium japonicum
Biochemistry, 45, 2006
|
|
1WUE
 
 | | Crystal structure of protein GI:29375081, unknown member of enolase superfamily from enterococcus faecalis V583 | | Descriptor: | mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-05 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1WUF
 
 | | Crystal structure of protein GI:16801725, member of Enolase superfamily from Listeria innocua Clip11262 | | Descriptor: | MAGNESIUM ION, hypothetical protein lin2664 | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1WHP
 
 | | ALLERGEN PHL P 2 | | Descriptor: | ALLERGEN PHL P 2 | | Authors: | Fedorov, A.A, Fedorov, E.V, Dolecek, C, Susani, M, Valenta, R, Almo, S.C. | | Deposit date: | 1997-04-04 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Phl P 2, a Major Timothy Grass (Phleum Pratense) Pollen Allergen
To be Published
|
|
1XM5
 
 | | Crystal structure of metal-dependent hydrolase ybeY from E. coli, Pfam UPF0054 | | Descriptor: | Hypothetical UPF0054 protein ybeY, NICKEL (II) ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Shi, W, Ramagopal, U.A, Thirumuruhan, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-10-01 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The ybeY protein from Escherichia coli is a metalloprotein.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1YS9
 
 | |
1YRZ
 
 | | Crystal structure of xylan beta-1,4-xylosidase from Bacillus Halodurans C-125 | | Descriptor: | xylan beta-1,4-xylosidase | | Authors: | Fedorov, A.A, Fedorov, E.V, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-02-05 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of xylan beta-1,4-xylosidase from Bacillus Halodurans C-125
To be Published
|
|
