1M2N
 
 | |
1M2H
 
 | | Sir2 homologue S24A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
7YQ8
 
 | | Cryo-EM structure of human topoisomerase II beta in complex with DNA and etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, 50-mer DNA, DNA topoisomerase 2-beta, ... | | Authors: | Naganuma, M, Ehara, H, Kim, D, Nakagawa, R, Cong, A, Bu, H, Jeong, J, Jang, J, Schellenberg, M.J, Bunch, H, Sekine, S. | | Deposit date: | 2022-08-05 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ERK2-topoisomerase II regulatory axis is important for gene activation in immediate early genes.
Nat Commun, 14, 2023
|
|
4N5M
 
 | |
4N5L
 
 | |
4N5N
 
 | |
6DF6
 
 | | Crystal structure of estrogen receptor alpha in complex with receptor degrader 16ab | | Descriptor: | (8R)-8-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-1,8-dihydro-2H-[1]benzopyrano[4,3-d][1]benzoxepine-5,11-diol, Estrogen receptor, GLYCEROL | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Ortwine, D.F, Nettles, K.W, Nwachukwu, J.C. | | Deposit date: | 2018-05-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unexpected equivalent potency of a constrained chromene enantiomeric pair rationalized by co-crystal structures in complex with estrogen receptor alpha.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6DFN
 
 | | Crystal structure of estrogen receptor alpha in complex with receptor degrader 16aa | | Descriptor: | (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, (8S)-8-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-1,8-dihydro-2H-[1]benzopyrano[4,3-d][1]benzoxepine-5,11-diol, Estrogen receptor, ... | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Ortwine, D.F, Nettles, K.W, Nwachukwu, J.C. | | Deposit date: | 2018-05-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected equivalent potency of a constrained chromene enantiomeric pair rationalized by co-crystal structures in complex with estrogen receptor alpha.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
5BVQ
 
 | | Ligand-unbound pFABP4 | | Descriptor: | fatty acid-binding protein | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
5BVT
 
 | | Palmitate-bound pFABP5 | | Descriptor: | Epidermal fatty acid-binding protein, PALMITOLEIC ACID | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
5BVS
 
 | | Linoleate-bound pFABP4 | | Descriptor: | Fatty acid-binding protein, LINOLEIC ACID | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
7C4X
 
 | |
6PET
 
 | | Crystal structure of 8-hydroxychromene compound 30 bound to estrogen receptor alpha | | Descriptor: | (2S)-2-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-3-(3-hydroxyphenyl)-4-methyl-2H-1-benzopyran-8-ol, (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, CHLORIDE ION, ... | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Wang, X, Zbieg, J, Labadie, S.S, Zhang, B, Li, J, Liang, W. | | Deposit date: | 2019-06-20 | | Release date: | 2019-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Discovery of a C-8 hydroxychromene as a potent degrader of estrogen receptor alpha with improved rat oral exposure over GDC-0927.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6PFM
 
 | | Crystal structure of GDC-0927 bound to estrogen receptor alpha | | Descriptor: | (2S)-2-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-3-(3-hydroxyphenyl)-4-methyl-2H-1-benzopyran-6-ol, Estrogen receptor | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Wang, X, Zbieg, J.R, Labadie, S.S, Li, J, Ray, N.C, Ortwine, D. | | Deposit date: | 2019-06-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of a C-8 hydroxychromene as a potent degrader of estrogen receptor alpha with improved rat oral exposure over GDC-0927.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
7MSA
 
 | | GDC-9545 in complex with estrogen receptor alpha | | Descriptor: | (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, 3-[(1R,3R)-1-(2,6-difluoro-4-{[1-(3-fluoropropyl)azetidin-3-yl]amino}phenyl)-3-methyl-1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl]-2,2-difluoropropan-1-ol, Estrogen receptor | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zbieg, J.R, Wang, X, Ortwine, D.F. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | GDC-9545 (Giredestrant): A Potent and Orally Bioavailable Selective Estrogen Receptor Antagonist and Degrader with an Exceptional Preclinical Profile for ER+ Breast Cancer.
J.Med.Chem., 64, 2021
|
|
6TGB
 
 | | CryoEM structure of the binary DOCK2-ELMO1 complex | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
6TGC
 
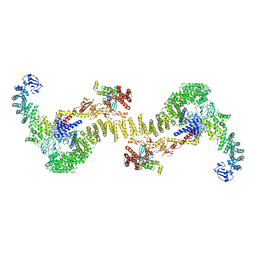 | | CryoEM structure of the ternary DOCK2-ELMO1-RAC1 complex. | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
6NO9
 
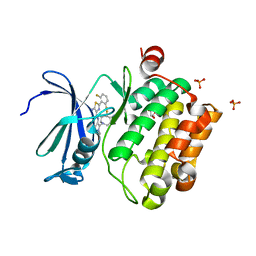 | | PIM1 in complex with Cpd16 (5-amino-N-(5-((4R,5R)-4-amino-5-fluoroazepan-1-yl)-1-methyl-1H-pyrazol-4-yl)-2-(2,6-difluorophenyl)thiazole-4-carboxamide) | | Descriptor: | 5-amino-N-{5-[(4R,5R)-4-amino-5-fluoroazepan-1-yl]-1-methyl-1H-pyrazol-4-yl}-2-(2,6-difluorophenyl)-1,3-thiazole-4-carboxamide, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Murray, J.M, Noland, C. | | Deposit date: | 2019-01-15 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Optimization of Pan-Pim Kinase Activity and Oral Bioavailability Leading to Diaminopyrazole (GDC-0339) for the Treatment of Multiple Myeloma.
J. Med. Chem., 62, 2019
|
|
7JV8
 
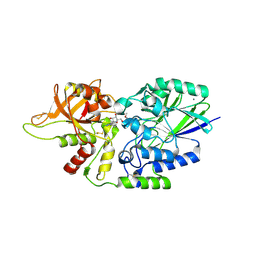 | | Human CD73 (ecto 5'-nucleotidase) in complex with compound 35 | | Descriptor: | 5'-nucleotidase, 6-chloro-N-cyclopentyl-1-{5-O-[(2R)-1-hydroxy-3-methoxy-2-phosphonopropan-2-yl]-beta-D-ribofuranosyl}-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALCIUM ION, ... | | Authors: | Gibbons, P, Du, X. | | Deposit date: | 2020-08-20 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Orally Bioavailable Small-Molecule CD73 Inhibitor (OP-5244) Reverses Immunosuppression through Blockade of Adenosine Production.
J.Med.Chem., 63, 2020
|
|
7JV9
 
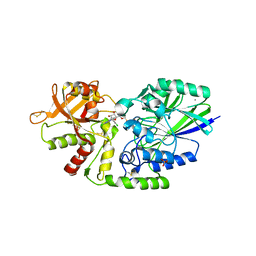 | | Human CD73 (ecto 5'-nucleotidase) in complex with compound 12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-nucleotidase, 6-chloro-N-[(2-chlorophenyl)methyl]-1-[5-O-(phosphonomethyl)-beta-D-ribofuranosyl]-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ... | | Authors: | Gibbons, P, Du, X. | | Deposit date: | 2020-08-20 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Orally Bioavailable Small-Molecule CD73 Inhibitor (OP-5244) Reverses Immunosuppression through Blockade of Adenosine Production.
J.Med.Chem., 63, 2020
|
|
5V80
 
 | |
5V82
 
 | | PIM1 kinase in complex with Cpd17 (1-(6-(4,4-difluoropiperidin-3-yl)pyridin-2-yl)-6-(6-methylpyrazin-2-yl)-1H-pyrazolo[4,3-c]pyridine) | | Descriptor: | 1-{6-[(3R)-4,4-difluoropiperidin-3-yl]pyridin-2-yl}-6-(6-methylpyrazin-2-yl)-1H-pyrazolo[4,3-c]pyridine, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Discovery of 5-Azaindazole (GNE-955) as a Potent Pan-Pim Inhibitor with Optimized Bioavailability.
J. Med. Chem., 60, 2017
|
|
4R1N
 
 | |
5DGZ
 
 | | Discovery of 3,5-substituted 6-azaindazoles as potent pan-Pim inhibitors | | Descriptor: | (2S)-1-(1H-INDOL-3-YL)-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, PHOSPHATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H, Steffek, M. | | Deposit date: | 2015-08-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Discovery of 3,5-substituted 6-azaindazoles as potent pan-Pim inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5DHJ
 
 | | PIM1 in complex with Cpd4 (3-methyl-5-(pyridin-3-yl)-1H-pyrazolo[3,4-c]pyridine) | | Descriptor: | 3-methyl-5-(pyridin-3-yl)-2H-pyrazolo[3,4-c]pyridine, PHOSPHATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H. | | Deposit date: | 2015-08-31 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.457 Å) | | Cite: | Discovery of 3,5-substituted 6-azaindazoles as potent pan-Pim inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
