7WJB
 
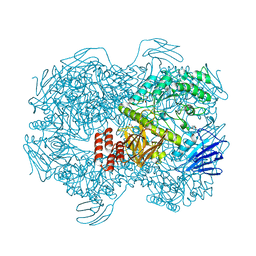 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose, ... | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJE
 
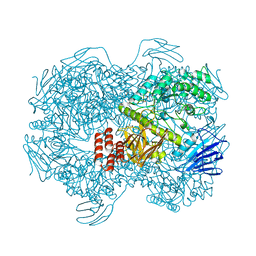 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with nigerotetraose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJ9
 
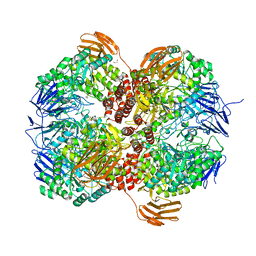 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase, P21 space group | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase, Xylitol | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WJD
 
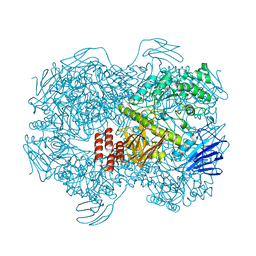 | | Crystal structure of Lactococcus lactis subsp. cremoris GH31 alpha-1,3-glucosidase mutant D394A in complex with nigerotriose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-xylosidase, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose, ... | | Authors: | Ikegaya, M, Miyazaki, T. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7XI9
 
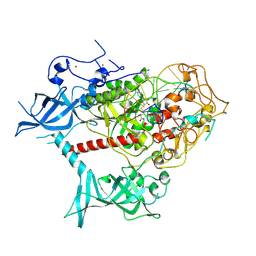 | | Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-ordered form) | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*TP*CP*(C55)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Onoda, H, Kikuchi, A, Kori, S, Yoshimi, S, Yamagata, A, Arita, K. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural basis for activation of DNMT1.
Nat Commun, 13, 2022
|
|
7XIB
 
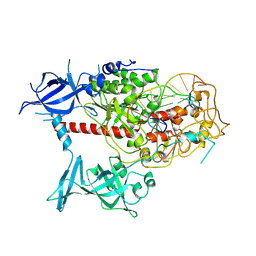 | | Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-disordered form) | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*TP*CP*(C55)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Onoda, H, Kikuchi, A, Kori, S, Yoshimi, S, Yamagata, A, Arita, K. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Structural basis for activation of DNMT1.
Nat Commun, 13, 2022
|
|
2E50
 
 | | Crystal structure of SET/TAF-1beta/INHAT | | Descriptor: | Protein SET, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Muto, S, Senda, M, Senda, T, Horikoshi, M. | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
