7POQ
 
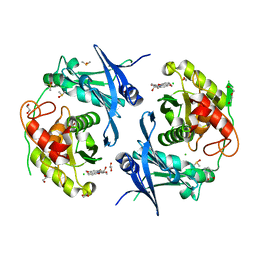 | | Crystal structure of profragilysin-3 (proBFT-3) from Bacteroides fragilis in complex with foliosidine in P41212. | | Descriptor: | 1,2-ETHANEDIOL, BFT-3, CHLORIDE ION, ... | | Authors: | Eckhard, U, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis.
Protein Sci., 31, 2022
|
|
7POU
 
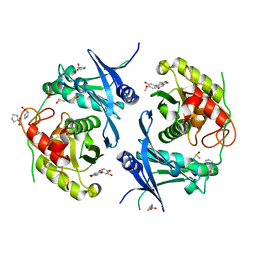 | | Crystal structure of profragilysin-3 (proBFT-3) from Bacteroides fragilis in complex with hesperetin. | | Descriptor: | (2S)-5,7-dihydroxy-2-(3-hydroxy-4-methoxyphenyl)-2,3-dihydro-4H-1-benzopyran-4-one, BFT-3, DIMETHYL SULFOXIDE, ... | | Authors: | Eckhard, U, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis.
Protein Sci., 31, 2022
|
|
7POO
 
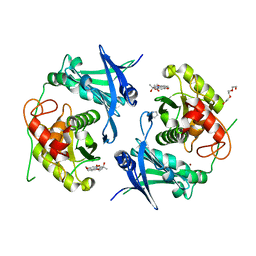 | | Crystal structure of profragilysin-3 (proBFT-3) from Bacteroides fragilis in complex with foliosidine in P212121. | | Descriptor: | ACETATE ION, BFT-3, PROLINE, ... | | Authors: | Eckhard, U, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis.
Protein Sci., 31, 2022
|
|
7PND
 
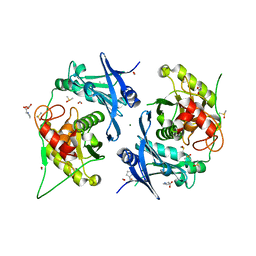 | | Crystal structure of profragilysin-3 (proBFT-3) from Bacteroides fragilis at 1.85 A resolution. | | Descriptor: | BFT-3, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Eckhard, U, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-09-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis.
Protein Sci., 31, 2022
|
|
7POL
 
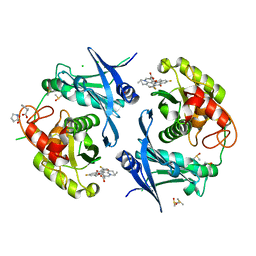 | | Crystal structure of profragilysin-3 (proBFT-3) from Bacteroides fragilis in complex with flumequine | | Descriptor: | (12~{R})-7-fluoranyl-12-methyl-4-oxidanylidene-1-azatricyclo[7.3.1.0^{5,13}]trideca-2,5(13),6,8-tetraene-3-carboxylic acid, BFT-3, CHLORIDE ION, ... | | Authors: | Eckhard, U, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis.
Protein Sci., 31, 2022
|
|
3H8T
 
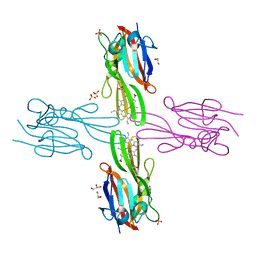 | | Structure of Porphyromonas gingivalis heme-binding protein HmuY in complex with Heme | | Descriptor: | GLYCEROL, HmuY, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wojtowicz, H, Guevara, T, Tallant, C, Olczak, M, Sroka, A, Potempa, J, Sola, M, Olczak, T, Gomis-Ruth, F.X. | | Deposit date: | 2009-04-29 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unique structure and stability of HmuY, a novel heme-binding protein of Porphyromonas gingivalis
Plos Pathog., 5, 2009
|
|
5MUN
 
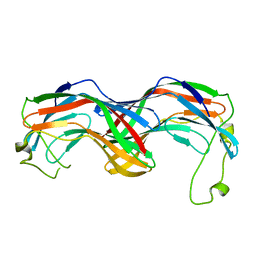 | | Structural insight into zymogenic latency of gingipain K from Porphyromonas gingivalis. | | Descriptor: | AZIDE ION, Lys-gingipain W83 | | Authors: | Pomowski, A, Uson, I, Nowakovska, Z, Veillard, F, Sztukowska, M.N, Guevara, T, Goulas, T, Mizgalska, D, Nowak, M, Potempa, B, Huntington, J.A, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights unravel the zymogenic mechanism of the virulence factor gingipain K from Porphyromonas gingivalis, a causative agent of gum disease from the human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
5NV6
 
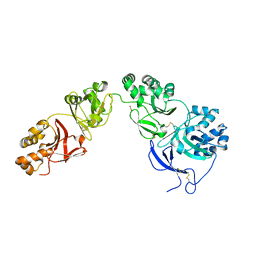 | | Structure of human transforming growth factor beta-induced protein (TGFBIp). | | Descriptor: | ACETATE ION, Transforming growth factor-beta-induced protein ig-h3 | | Authors: | Garcia-Castellanos, R, Nielsen, S.N, Runager, K, Thogersen, B.I, Goulas, T, Enghild, J.J, Gomis-Ruth, F.X. | | Deposit date: | 2017-05-03 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural and Functional Implications of Human Transforming Growth Factor beta-Induced Protein, TGFBIp, in Corneal Dystrophies.
Structure, 25, 2017
|
|
3LUM
 
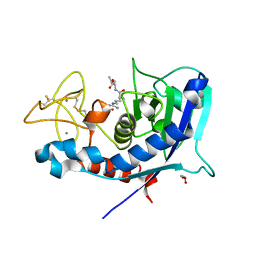 | | Structure of ulilysin mutant M290L | | Descriptor: | ARGININE, CALCIUM ION, GLYCEROL, ... | | Authors: | Tallant, C, Garcia-Castellanos, R, Baumann, U, Gomis-Ruth, F.X. | | Deposit date: | 2010-02-18 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | On the relevance of the Met-turn methionine in metzincins.
J.Biol.Chem., 285, 2010
|
|
3LQ0
 
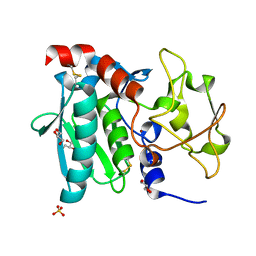 | | Zymogen structure of crayfish astacin metallopeptidase | | Descriptor: | GLYCEROL, ProAstacin, SULFATE ION, ... | | Authors: | Guevara, T, Yiallouros, I, Kappelhoff, R, Bissdorf, S, Stocker, W, Gomis-Ruth, F.X. | | Deposit date: | 2010-02-08 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Proenzyme structure and activation of astacin metallopeptidase
J.Biol.Chem., 285, 2010
|
|
3LUN
 
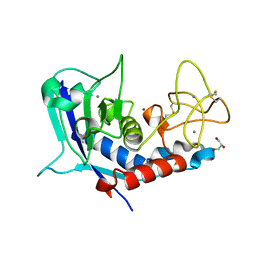 | | Structure of ulilysin mutant M290C | | Descriptor: | ARGININE, CALCIUM ION, GLYCEROL, ... | | Authors: | Tallant, C, Garcia-Castellanos, R, Baumann, U, Gomis-Ruth, F.X. | | Deposit date: | 2010-02-18 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | On the relevance of the Met-turn methionine in metzincins.
J.Biol.Chem., 285, 2010
|
|
3DKX
 
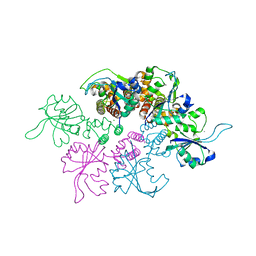 | | Crystal Structure of the replication initiator protein encoded on plasmid pMV158 (RepB), trigonal form, to 2.7 Ang resolution | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Boer, D.R, Ruiz-Maso, J.A, Blanco, A.G, Vives-Llacer, M, Uson, I, Gomis-Ruth, F.X, Espinosa, M, Del Solar, G, Coll, M. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains
Embo J., 28, 2009
|
|
3DKY
 
 | | Crystal Structure of the replication initiator protein encoded on plasmid pMV158 (RepB), tetragonal form, to 3.6 Ang resolution | | Descriptor: | MANGANESE (II) ION, Replication protein repB | | Authors: | Boer, D.R, Ruiz-Maso, J.A, Blanco, A.G, Vives-Llacer, M, Uson, I, Gomis-Ruth, F.X, Espinosa, M, Del Solar, G, Coll, M. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains
Embo J., 28, 2009
|
|
2J83
 
 | | Ulilysin metalloprotease in complex with batimastat. | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, GLYCEROL, ... | | Authors: | Garcia-Castellanos, R, Tallant, C, Marrero, A, Sola, M, Baumann, U, Gomis-Ruth, F.X. | | Deposit date: | 2006-10-18 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Specificity of a Metalloprotease of the Pappalysin Family Revealed by an Inhibitor and a Product Complex.
Arch.Biochem.Biophys., 457, 2007
|
|
2JB9
 
 | | PhoB response regulator receiver domain constitutively-active double mutant D10A and D53E. | | Descriptor: | PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | Authors: | Ferrer-Orta, C, Arribas-Bosacoma, R, Kim, S.-K, Blanco, A.G, Pereira, P.J.B, Gomis-Ruth, F.X, Wanner, B.L, Coll, M, Sola, M. | | Deposit date: | 2006-12-05 | | Release date: | 2007-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The X-Ray Crystal Structures of Two Constitutively Active Mutants of the E. Coli Phob Receiver Domain Give Insights Into Activation
J.Mol.Biol., 366, 2007
|
|
2JBA
 
 | | PhoB response regulator receiver domain constitutively-active double mutant D53A and Y102C. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB, SODIUM ION | | Authors: | Arribas-Bosacoma, R, Ferrer-Orta, C, Kim, S.-K, Blanco, A.G, Pereira, P.J.B, Gomis-Ruth, F.X, Wanner, B.L, Coll, M, Sola, M. | | Deposit date: | 2006-12-05 | | Release date: | 2007-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The X-Ray Crystal Structures of Two Constitutively Active Mutants of the Escherichia Coli Phob Receiver Domain Give Insights Into Activation.
J.Mol.Biol., 366, 2007
|
|
7SKN
 
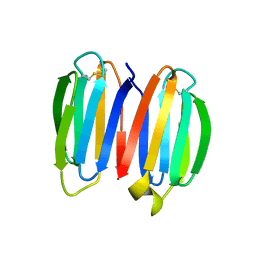 | |
7SKO
 
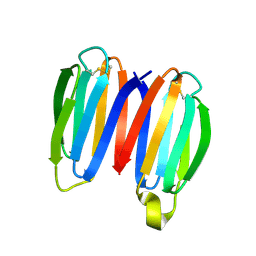 | | De novo synthetic protein DIG8-CC (orthogonal space group) | | Descriptor: | De novo synthetic protein DIG8-CC, MAGNESIUM ION | | Authors: | Mendes, S.R, Eckhard, U, Marcos, E, Gomis-Ruth, F.X. | | Deposit date: | 2021-10-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | De novo design of immunoglobulin-like domains
Nat Commun, 13, 2022
|
|
7SKP
 
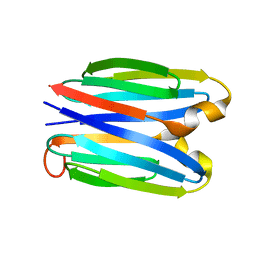 | |
7O7O
 
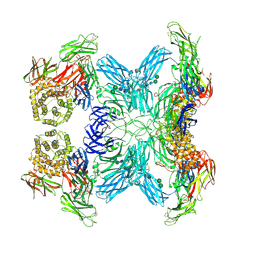 | | (h-alpha2M)4 semiactivated II state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin, ... | | Authors: | Luque, D, Goulas, T, Mata, C.P, Mendes, S.R, Gomis-Ruth, F.X, Caston, J.R. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structures show the mechanistic basis of pan-peptidase inhibition by human alpha 2 -macroglobulin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7O7S
 
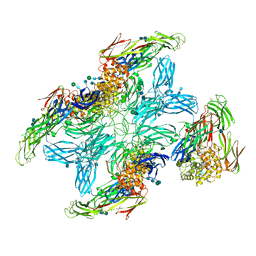 | | (h-alpha2M)4 plasmin-activated II state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin, ... | | Authors: | Luque, D, Goulas, T, Mata, C.P, Mendes, S.R, Gomis-Ruth, F.X, Caston, J.R. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures show the mechanistic basis of pan-peptidase inhibition by human alpha 2 -macroglobulin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7O7R
 
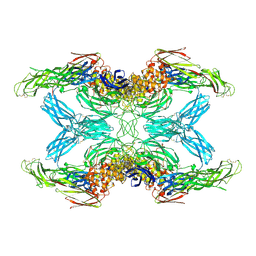 | | (h-alpha2M)4 plasmin-activated I state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin, ... | | Authors: | Luque, D, Goulas, T, Mata, C.P, Mendes, S.R, Gomis-Ruth, F.X, Caston, J.R. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures show the mechanistic basis of pan-peptidase inhibition by human alpha 2 -macroglobulin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7O7M
 
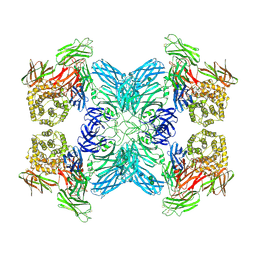 | | (h-alpha2M)4 native II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin, ... | | Authors: | Luque, D, Goulas, T, Mata, C.P, Mendes, S.R, Gomis-Ruth, F.X, Caston, J.R. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structures show the mechanistic basis of pan-peptidase inhibition by human alpha 2 -macroglobulin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7O7N
 
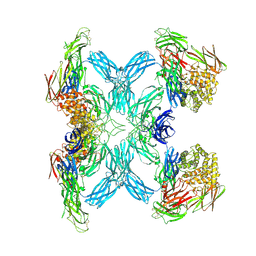 | | (h-alpha2M)4 semiactivated I state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin | | Authors: | Luque, D, Goulas, T, Mata, C.P, Mendes, S.R, Gomis-Ruth, F.X, Caston, J.R. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Cryo-EM structures show the mechanistic basis of pan-peptidase inhibition by human alpha 2 -macroglobulin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7O7Q
 
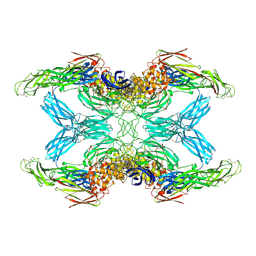 | | (h-alpha2M)4 trypsin-activated state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-2-macroglobulin, ... | | Authors: | Luque, D, Goulas, T, Mata, C.P, Mendes, S.R, Gomis-Ruth, F.X, Caston, J.R. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures show the mechanistic basis of pan-peptidase inhibition by human alpha 2 -macroglobulin.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
