8X6B
 
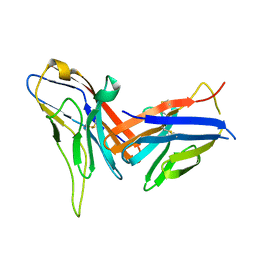 | | Crystal structure of immune receptor PVRIG in complex with ligand Nectin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nectin-2, Transmembrane protein PVRIG | | Authors: | Hu, S.T, Han, P, Wang, H, Qi, J.X. | | Deposit date: | 2023-11-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the immune recognition and selectivity of the immune receptor PVRIG for ligand Nectin-2.
Structure, 2024
|
|
5XQ1
 
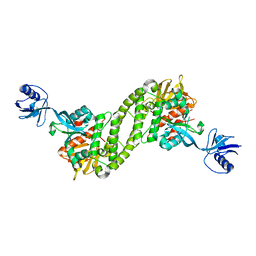 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2,Integrin beta-3 | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6L4R
 
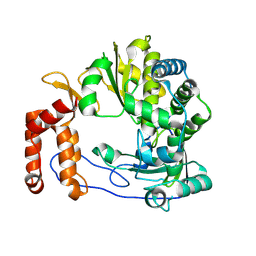 | | Crystal structure of Enterovirus D68 RdRp | | Descriptor: | RdRp | | Authors: | Wang, M.L, Li, L, Zhang, Y, Chen, Y.P, Su, D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 211, 2020
|
|
4F23
 
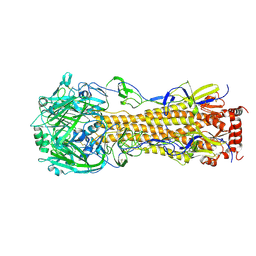 | | Influenza A virus hemagglutinin H16 HA0 structure with an alpha-helix conformation in the cleavage site: a potential drug target | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Lu, X, Shi, Y, Gao, F, Xiao, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-07 | | Release date: | 2012-11-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into Avian Influenza Virus Pathogenicity: the Hemagglutinin Precursor HA0 of Subtype H16 Has an Alpha-Helix Structure in Its Cleavage Site with Inefficient HA1/HA2 Cleavage.
J.Virol., 86, 2012
|
|
8WYP
 
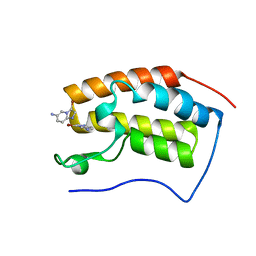 | |
4U68
 
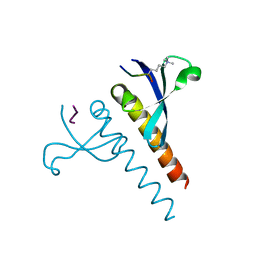 | |
8EF5
 
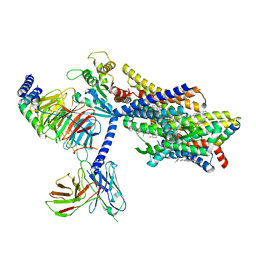 | | Fentanyl-bound mu-opioid receptor-Gi complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EFB
 
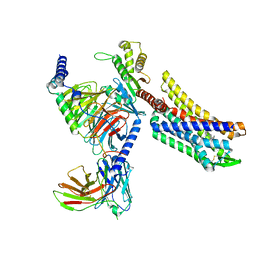 | | Oliceridine-bound mu-opioid receptor-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EF6
 
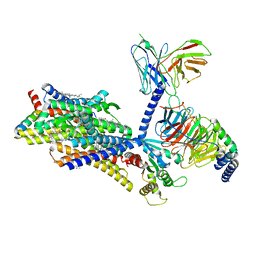 | | Morphine-bound mu-opioid receptor-Gi complex | | Descriptor: | (7R,7AS,12BS)-3-METHYL-2,3,4,4A,7,7A-HEXAHYDRO-1H-4,12-METHANO[1]BENZOFURO[3,2-E]ISOQUINOLINE-7,9-DIOL, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EFQ
 
 | | DAMGO-bound mu-opioid receptor-Gi complex | | Descriptor: | DAMGO, ETHANOLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EFO
 
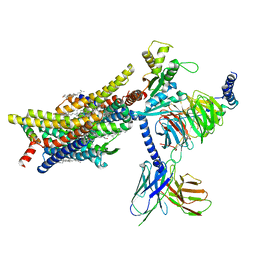 | | PZM21-bound mu-opioid receptor-Gi complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EFL
 
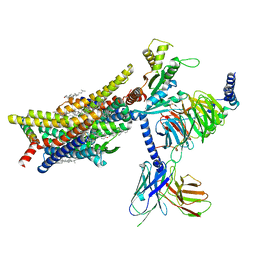 | | SR17018-bound mu-opioid receptor-Gi complex | | Descriptor: | 5,6-dichloro-1-{1-[(4-chlorophenyl)methyl]piperidin-4-yl}-1,3-dihydro-2H-benzimidazol-2-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
1SN4
 
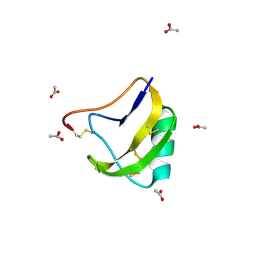 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M4 | | Descriptor: | ACETATE ION, PROTEIN (NEUROTOXIN BMK M4) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
8T5C
 
 | | Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8.11G Heavy Chain, 8.11G Light Chain, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cleavage-intermediate Lassa virus trimer elicits neutralizing responses, identifies neutralizing nanobodies, and reveals an apex-situated site-of-vulnerability.
Nat Commun, 15, 2024
|
|
6M53
 
 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, GLYCEROL, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
6LYW
 
 | | Structural insight into the biological functions of Arabidopsis thaliana ACHT1 | | Descriptor: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | Authors: | Wang, J.C, Pan, W.M, Wang, M.Z, Zhang, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
6LYX
 
 | | Crystal structure of oxidized ACHT1 | | Descriptor: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | Authors: | Wang, J.C, Pan, W.M, Cai, W.G, Wang, M.Z, Zhang, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
6U3J
 
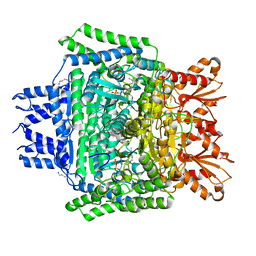 | | Structure of the 2-oxoadipate dehydrogenase DHTKD1 | | Descriptor: | 2-oxoglutarate dehydrogenase E1 component DHKTD1, mitochondrial, MAGNESIUM ION, ... | | Authors: | Khamrui, S, Lazarus, M.B. | | Deposit date: | 2019-08-21 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Inhibition and Crystal Structure of the Human DHTKD1-Thiamin Diphosphate Complex.
Acs Chem.Biol., 15, 2020
|
|
4TZU
 
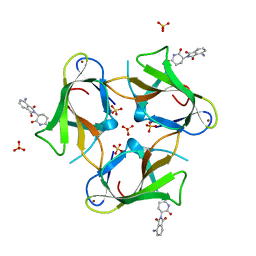 | | Crystal Structure of Murine Cereblon in Complex with Pomalidomide | | Descriptor: | Protein cereblon, S-Pomalidomide, SULFATE ION, ... | | Authors: | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
to be published
|
|
4TZC
 
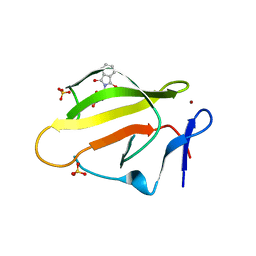 | | Crystal Structure of Murine Cereblon in Complex with Thalidomide | | Descriptor: | Protein cereblon, S-Thalidomide, SULFATE ION, ... | | Authors: | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
to be published
|
|
4U93
 
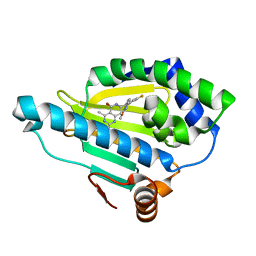 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | Descriptor: | (7R)-2-amino-7-[4-fluoro-2-(6-methoxypyridin-2-yl)phenyl]-4-methyl-7,8-dihydropyrido[4,3-d]pyrimidin-5(6H)-one, Heat shock protein HSP 90-alpha | | Authors: | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | Deposit date: | 2014-08-05 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
4W7T
 
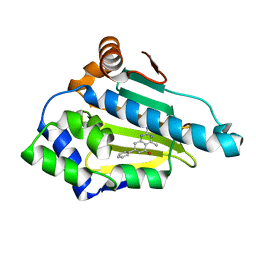 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | Descriptor: | (7S)-2-amino-4-methyl-7-phenyl-7,8-dihydroquinazolin-5(6H)-one, Heat shock protein HSP 90-alpha | | Authors: | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | Deposit date: | 2014-08-22 | | Release date: | 2014-11-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
1SN1
 
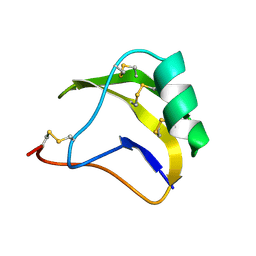 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M1 | | Descriptor: | PROTEIN (NEUROTOXIN BMK M1) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
5PGU
 
 | |
5PGX
 
 | |
