3Q4J
 
 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
1BHX
 
 | |
3IW4
 
 | | Crystal structure of PKC alpha in complex with NVP-AEB071 | | Descriptor: | 3-(1H-indol-3-yl)-4-[2-(4-methylpiperazin-1-yl)quinazolin-4-yl]-1H-pyrrole-2,5-dione, Protein kinase C alpha type | | Authors: | Stark, W, Rummel, G, Strauss, A, Cowan-Jacob, S.W. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 3-(1H-indol-3-yl)-4-[2-(4-methylpiperazin-1-yl)quinazolin-4-yl]pyrrole-2,5-dione (AEB071), a potent and selective inhibitor of protein kinase C isotypes
J.Med.Chem., 52, 2009
|
|
5FBE
 
 | | COMPLEMENT FACTOR D IN COMPLEX WITH COMPOUND2 | | Descriptor: | Complement factor D, GLYCEROL, methyl 2-[[[(2~{S})-2-[[3-(trifluoromethyloxy)phenyl]carbamoyl]pyrrolidin-1-yl]carbonylamino]methyl]benzoate | | Authors: | Ostermann, N, Zink, F. | | Deposit date: | 2015-12-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
5FAH
 
 | | KALLIKREIN-7 IN COMPLEX WITH COMPOUND1 | | Descriptor: | (2~{S})-~{N}2-[2-(4-methoxyphenyl)ethyl]-~{N}1-(naphthalen-1-ylmethyl)pyrrolidine-1,2-dicarboxamide, ACETATE ION, Kallikrein-7 | | Authors: | Ostermann, N, Zink, F. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
5FCK
 
 | | COMPLEMENT FACTOR D IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 1-[2-[(1~{R},3~{S},5~{R})-3-[[(1~{R})-1-(3-chloranyl-2-fluoranyl-phenyl)ethyl]carbamoyl]-2-azabicyclo[3.1.0]hexan-2-yl]-2-oxidanylidene-ethyl]pyrazolo[3,4-c]pyridine-3-carboxamide, Complement factor D, SULFATE ION | | Authors: | Mac Sweeney, A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
5FBI
 
 | | COMPLEMENT FACTOR D IN COMPLEX WITH COMPOUND 3b | | Descriptor: | 3-[(2-aminocarbonyl-1~{H}-indol-5-yl)oxymethyl]benzoic acid, Complement factor D, GLYCEROL | | Authors: | Ostermann, N, Zink, F. | | Deposit date: | 2015-12-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
3TUU
 
 | | Structure of dihydrodipicolinate synthase from the common grapevine | | Descriptor: | BROMIDE ION, CHLORIDE ION, dihydrodipicolinate synthase | | Authors: | Perugini, M.A, Dobson, R.C, Atkinson, S.C. | | Deposit date: | 2011-09-19 | | Release date: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal, Solution and In silico Structural Studies of Dihydrodipicolinate Synthase from the Common Grapevine.
Plos One, 7, 2012
|
|
8GCC
 
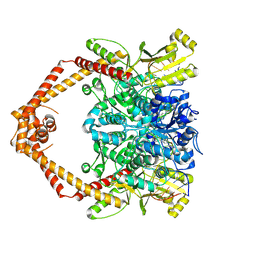 | | T. cruzi topoisomerase II alpha bound to dsDNA and the covalent inhibitor CT1 | | Descriptor: | 2-{3-[(Z)-iminomethyl]-1H-1,2,4-triazol-1-yl}-1-{(3M)-3-[2-(trifluoromethyl)phenyl]-6H-pyrrolo[3,4-b]pyridin-6-yl}ethan-1-one, DNA (28-MER), DNA topoisomerase 2 | | Authors: | Schenk, A, Deniston, C, Noeske, J. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cyanotriazoles are selective topoisomerase II poisons that rapidly cure trypanosome infections.
Science, 380, 2023
|
|
2FPU
 
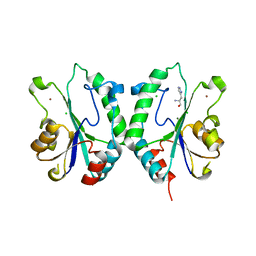 | | Crystal Structure of the N-terminal domain of E.coli HisB- Complex with histidinol | | Descriptor: | CHLORIDE ION, Histidine biosynthesis bifunctional protein hisB, L-histidinol, ... | | Authors: | Rangarajan, E.S, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway.
J.Biol.Chem., 281, 2006
|
|
2FPX
 
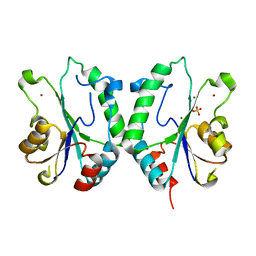 | | Crystal Structure of the N-terminal Domain of E.coli HisB- Sulfate complex. | | Descriptor: | Histidine biosynthesis bifunctional protein hisB, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Rangarajan, E.S, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway.
J.Biol.Chem., 281, 2006
|
|
2FPW
 
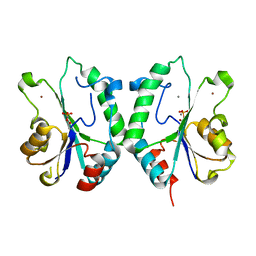 | | Crystal Structure of the N-terminal Domain of E.coli HisB- Phosphoaspartate intermediate. | | Descriptor: | CALCIUM ION, Histidine biosynthesis bifunctional protein hisB, ZINC ION | | Authors: | Rangarajan, E.S, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway.
J.Biol.Chem., 281, 2006
|
|
2FPR
 
 | | Crystal structure the N-terminal domain of E. coli HisB. Apo Mg model. | | Descriptor: | BROMIDE ION, Histidine biosynthesis bifunctional protein hisB, MAGNESIUM ION, ... | | Authors: | Rangarajan, E.S, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-17 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway.
J.Biol.Chem., 281, 2006
|
|
2FPS
 
 | | Crystal structure of the N-terminal domain of E.coli HisB- Apo Ca model. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Histidine biosynthesis bifunctional protein hisB, ... | | Authors: | Rangarajan, E.S, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway.
J.Biol.Chem., 281, 2006
|
|
6Q7A
 
 | |
6Q6O
 
 | |
6Q7H
 
 | |
6Q6M
 
 | |
8PAT
 
 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 3. | | Descriptor: | ACE-GLN-ALC-GLX-LEU-PHE, Beta sliding clamp | | Authors: | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, Guichard, G. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
5FCR
 
 | | MOUSE COMPLEMENT FACTOR D | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Complement factor D, DIMETHYL SULFOXIDE, ... | | Authors: | Mac Sweeney, A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
4TR6
 
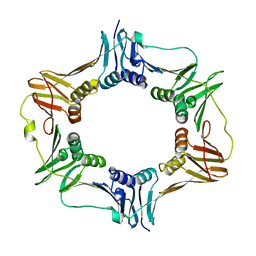 | | Crystal structure of DNA polymerase sliding clamp from Bacillus subtilis | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION | | Authors: | Burnouf, D, Olieric, V, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-14 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
4TSZ
 
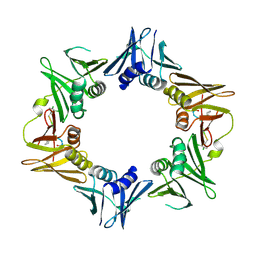 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa with ligand | | Descriptor: | ACE-GLN-ALC-ASP-LEU-ZCL peptide, DNA polymerase III subunit beta | | Authors: | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-19 | | Release date: | 2014-09-10 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
3CES
 
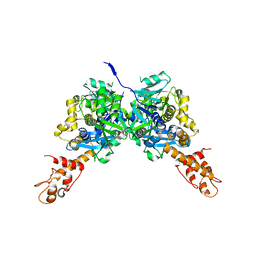 | | Crystal Structure of E.coli MnmG (GidA), a Highly-Conserved tRNA Modifying Enzyme | | Descriptor: | tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2008-02-29 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme.
J.Bacteriol., 191, 2009
|
|
3G05
 
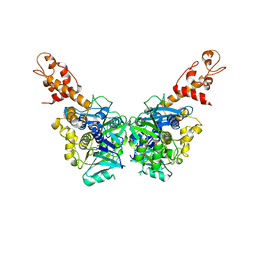 | | Crystal structure of N-terminal domain (2-550) of E.coli MnmG | | Descriptor: | SULFATE ION, tRNA uridine 5-carboxymethylaminomethyl modification enzyme mnmG | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2009-01-27 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme.
J.Bacteriol., 191, 2009
|
|
6G07
 
 | | RORGT (264-518;C455S) IN COMPLEX WITH INVERSE AGONIST "CPD-9" AND RIP140 PEPTIDE AT 1.66A | | Descriptor: | Nuclear receptor ROR-gamma, Nuclear receptor-interacting protein 1, ~{N}-[5-chloranyl-6-[(1~{S})-1-phenylethoxy]pyridin-3-yl]-2-(4-ethylsulfonylphenyl)ethanamide | | Authors: | Kallen, J. | | Deposit date: | 2018-03-16 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Optimizing a Weakly Binding Fragment into a Potent ROR gamma t Inverse Agonist with Efficacy in an in Vivo Inflammation Model.
J. Med. Chem., 61, 2018
|
|
