6EE7
 
 | | Small tetraheme cytochrome c from Shewanella oneidensis | | Descriptor: | HEME C, Periplasmic tetraheme cytochrome c CctA, ZINC ION | | Authors: | Huang, J, Zarzycki, J, Ducat, D.C, Kramer, D.M. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.394 Å) | | Cite: | Mesoscopic to Macroscopic Electron Transfer by Hopping in a Crystal Network of Cytochromes.
J.Am.Chem.Soc., 142, 2020
|
|
8AFV
 
 | | DaArgC3 - Engineered Formyl Phosphate Reductase with 3 substitutions (S178V, G182V, L233I) | | Descriptor: | N-acetyl-gamma-glutamyl-phosphate reductase, SODIUM ION | | Authors: | Pfister, P, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2022-07-18 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | N-acetyl-gamma-glutamyl-phosphate reductase of Denitrovibrio acetiphilus
To Be Published
|
|
8AFU
 
 | |
8APQ
 
 | | CaMct - Mesaconyl-CoA C1:C4 CoA Transferase of Chloroflexus aurantiacus | | Descriptor: | (2E)-2-METHYLBUT-2-ENEDIOIC ACID, 2-methylfumaryl-CoA isomerase, COENZYME A, ... | | Authors: | Pfister, P, Zarzycki, J, Erb, T.J. | | Deposit date: | 2022-08-10 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Basis for a Cork-Up Mechanism of the Intra-Molecular Mesaconyl-CoA Transferase.
Biochemistry, 62, 2023
|
|
8APR
 
 | |
8OIE
 
 | | Iron Nitrogenase Complex from Rhodobacter capsulatus | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Schmidt, F.V, Schulz, L, Zarzycki, J, Prinz, S, Erb, T.J, Rebelein, J.G. | | Deposit date: | 2023-03-22 | | Release date: | 2023-10-04 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural insights into the iron nitrogenase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PBB
 
 | | CHAPSO treated partial catalytic component (comprising only AnfD & AnfK, lacking AnfG and FeFeco) of iron nitrogenase from Rhodobacter capsulatus | | Descriptor: | FE(8)-S(7) CLUSTER, Nitrogenase iron-iron protein, beta subunit, ... | | Authors: | Schmidt, F.V, Schulz, L, Zarzycki, J, Prinz, S, Erb, T.J, Rebelein, J.G. | | Deposit date: | 2023-06-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural insights into the iron nitrogenase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7QVI
 
 | | Fiber-forming RubisCO derived from ancestral sequence reconstruction and rational engineering | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | Authors: | Schulz, L, Zarzycki, J, Prinz, S, Schuller, J.M, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
6OWE
 
 | | Enoyl-CoA carboxylases/reductases in complex with ethylmalonyl CoA | | Descriptor: | 5'-O-[(S)-{[(S)-[(3R)-4-({(1E)-3-[(2-{[(2S)-2-carboxybutanoyl]sulfanyl}ethyl)amino]-3-oxoprop-1-en-1-yl}amino)-3-hydroxy-2,2-dimethyl-4-oxobutoxy](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]adenosine 3'-(dihydrogen phosphate), Crotonyl-CoA carboxylase/reductase, IMIDAZOLE, ... | | Authors: | DeMirci, H. | | Deposit date: | 2019-05-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Four amino acids define the CO2binding pocket of enoyl-CoA carboxylases/reductases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5DIH
 
 | |
5DII
 
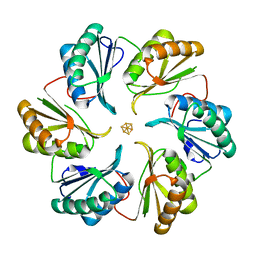 | | Structure of an engineered bacterial microcompartment shell protein binding a [4Fe-4S] cluster | | Descriptor: | IRON/SULFUR CLUSTER, Microcompartments protein | | Authors: | Sutter, M, Aussignargues, C, Turmo, A, Kerfeld, C.A. | | Deposit date: | 2015-09-01 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structure and Function of a Bacterial Microcompartment Shell Protein Engineered to Bind a [4Fe-4S] Cluster.
J.Am.Chem.Soc., 138, 2016
|
|
6SL8
 
 | | Diaminobutyrate acetyltransferase EctA from Paenibacillus lautus in complex with its substrate L-2,4-diaminobutyric acid (DAB) | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, GLYCEROL, L-2,4-diaminobutyric acid acetyltransferase, ... | | Authors: | Richter, A.A, Kobus, S, Czech, L, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine.
J.Biol.Chem., 295, 2020
|
|
6SK1
 
 | | Diaminobutyrate acetyltransferase EctA from Paenibacillus lautus in complex with coenzyme A | | Descriptor: | ACETATE ION, COENZYME A, L-2,4-diaminobutyric acid acetyltransferase | | Authors: | Richter, A.A, Kobus, S, Czech, L, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2019-08-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine.
J.Biol.Chem., 295, 2020
|
|
6SLL
 
 | | Diaminobutyrate acetyltransferase EctA from Paenibacillus lautus in complex with its substrate L-2,4-diaminobutyric acid (DAB) and coenzyme A | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, COENZYME A, L-2,4-diaminobutyric acid acetyltransferase, ... | | Authors: | Richter, A.A, Kobus, S, Czech, L, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2019-08-20 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine.
J.Biol.Chem., 295, 2020
|
|
6SJY
 
 | | Diaminobutyrate acetyltransferase EctA from Paenibacillus lautus in complex with its product ADABA | | Descriptor: | (2~{S})-4-acetamido-2-azanyl-butanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Richter, A.A, Kobus, S, Czech, L, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2019-08-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine.
J.Biol.Chem., 295, 2020
|
|
6SLK
 
 | | Diaminobutyrate acetyltransferase EctA from Paenibacillus lautus | | Descriptor: | L-2,4-diaminobutyric acid acetyltransferase, SODIUM ION, SULFATE ION | | Authors: | Richter, A.A, Kobus, S, Czech, L, Hoeppner, A, Bremer, E, Smits, S.H.J. | | Deposit date: | 2019-08-20 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine.
J.Biol.Chem., 295, 2020
|
|
5V76
 
 | | Structure of Haliangium ochraceum BMC-T HO-3341 | | Descriptor: | GLYCEROL, Microcompartments protein | | Authors: | Sutter, M, Paasch, B, Zarzycki, J, Aussignargues, C, Kerfeld, C.A. | | Deposit date: | 2017-03-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell.
Science, 356, 2017
|
|
