1TUT
 
 | | J4/5 Loop from the Candida albicans and Candida dubliniensis Group I Introns | | Descriptor: | 5'-R(*GP*AP*GP*GP*AP*AP*GP*GP*CP*GP*A)-3', 5'-R(*UP*CP*GP*UP*UP*AP*AP*UP*CP*UP*C)-3' | | Authors: | Znosko, B.M, Kennedy, S.D, Wille, P.C, Krugh, T.R, Turner, D.H. | | Deposit date: | 2004-06-25 | | Release date: | 2004-12-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Features and Thermodynamics of the J4/5 Loop from the Candida albicans and Candida dubliniensis Group I Introns.
Biochemistry, 43, 2004
|
|
1G3A
 
 | |
1GUC
 
 | |
2L8F
 
 | |
2O81
 
 | |
2O83
 
 | |
2LX1
 
 | |
2KY0
 
 | |
2KY1
 
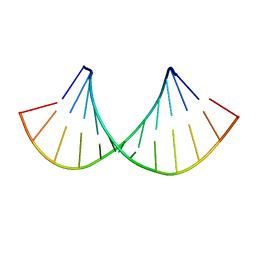 | |
2KY2
 
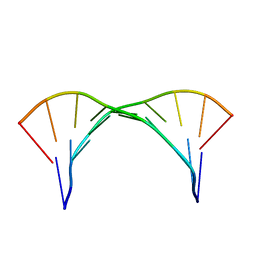 | |
2KXZ
 
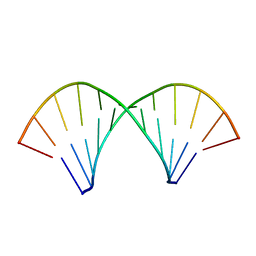 | |
1QET
 
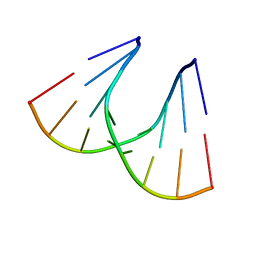 | | TANDEM GU MISMATCHES IN RNA, NMR, 30 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*AP*UP*GP*UP*CP*C)-3') | | Authors: | Mcdowell, J.A, He, L, Chen, X, Turner, D.H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGAUGUCC)2.
Biochemistry, 36, 1997
|
|
1QES
 
 | | TANDEM GU MISMATCHES IN RNA, NMR, 30 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*AP*GP*UP*UP*CP*C)-3') | | Authors: | Mcdowell, J.A, He, L, Chen, X, Turner, D.H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGAUGUCC)2.
Biochemistry, 36, 1997
|
|
1EKA
 
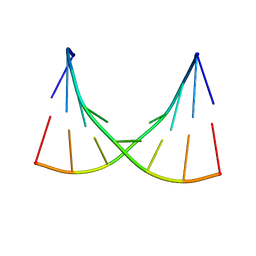 | | NMR AND MOLECULAR MODELING REVEAL THAT DIFFERENT HYDROGEN BONDING PATTERNS ARE POSSIBLE FOR GU PAIRS: ONE HYDROGEN BOND FOR EACH GU PAIR IN R(GGCGUGCC)2 AND TWO FOR EACH GU PAIR IN R(GAGUGCUC)2 | | Descriptor: | RNA (5'-R(*GP*AP*GP*UP*GP*CP*UP*C)-3') | | Authors: | Chen, X, McDowell, J.A, Kierzek, R, Krugh, T.R, Turner, D.H. | | Deposit date: | 2000-03-07 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2).
Biochemistry, 39, 2000
|
|
1EKD
 
 | | NMR AND MOLECULAR MODELING REVEAL THAT DIFFERENT HYDROGEN BONDING PATTERNS ARE POSSIBLE FOR GU PAIRS: ONE HYDROGEN BOND FOR EACH GU PAIR IN R(GGCGUGCC)2 AND TWO FOR EACH GU PAIR IN R(GAGUGCUC)2 | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*GP*CP*C)-3') | | Authors: | Chen, X, McDowell, J.A, Kierzek, R, Krugh, T.R, Turner, D.H. | | Deposit date: | 2000-03-07 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2).
Biochemistry, 39, 2000
|
|
5V2R
 
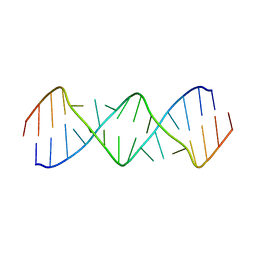 | | Structure of a GA Rich 8x8 Nucleotide RNA Internal Loop | | Descriptor: | RNA (5'-R(*CP*CP*AP*GP*AP*AP*AP*CP*GP*GP*AP*UP*GP*GP*A)-3') | | Authors: | Kauffmann, A.D, Kennedy, S.D, Turner, D.H. | | Deposit date: | 2017-03-06 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of an 8 8 Nucleotide RNA Internal Loop Flanked on Each Side by Three Watson-Crick Pairs and Comparison to Three-Dimensional Predictions.
Biochemistry, 56, 2017
|
|
5K8H
 
 | |
2JSE
 
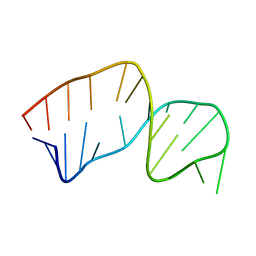 | |
2IRN
 
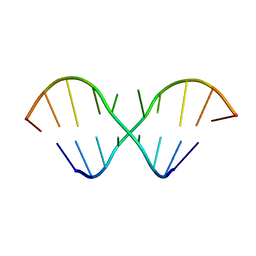 | | The NMR Structures of (rGCUGAGGCU)2 and (rGCGGAUGCU)2 | | Descriptor: | 5'-R(P*GP*CP*UP*GP*AP*GP*GP*CP*U)-3' | | Authors: | Tolbert, B.S. | | Deposit date: | 2006-10-15 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of (rGCUGAGGCU)(2) and (rGCGGAUGCU)(2): Probing the Structural Features That Shape the Thermodynamic Stability of GA Pairs(,).
Biochemistry, 46, 2007
|
|
2IRO
 
 | | The NMR Structures of (rGCUGAGGCU)2 and (rGCGGAUGCU)2 | | Descriptor: | 5'-R(P*GP*CP*GP*GP*AP*UP*GP*CP*U)-3' | | Authors: | Tolbert, B.S. | | Deposit date: | 2006-10-16 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of (rGCUGAGGCU)(2) and (rGCGGAUGCU)(2): Probing the Structural Features That Shape the Thermodynamic Stability of GA Pairs(,).
Biochemistry, 46, 2007
|
|
2H49
 
 | |
