8J8V
 
 | | Structure of beta-arrestin2 in complex with D6Rpp (Local Refine) | | Descriptor: | Atypical chemokine receptor 2, Beta-arrestin-2, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8J9K
 
 | | Structure of basal beta-arrestin2 | | Descriptor: | Beta-arrestin-2, Fab6 heavy chain, Fab6 light chain | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8IYW
 
 | | Structure of GSK256073-GPR109A-G-protein complex | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8IY9
 
 | | Structure of Niacin-GPR109A-G protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8JER
 
 | | Structure of Acipimox-GPR109A-G protein complex | | Descriptor: | 5-methyl-4-oxidanyl-pyrazin-4-ium-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8IYH
 
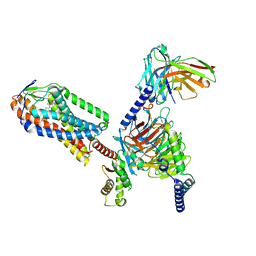 | | Structure of MK6892-GPR109A-G-protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8JHN
 
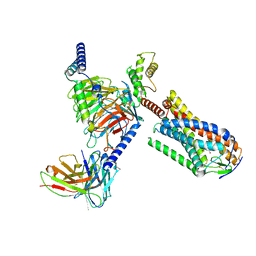 | | Structure of MMF-GPR109A-G protein complex | | Descriptor: | (E)-4-methoxy-4-oxidanylidene-but-2-enoic acid, G protein subunit alpha o1,Guanine nucleotide-binding protein G(o) subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-24 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
6F3V
 
 | | Backbone structure of bradykinin (BK) peptide bound to human Bradykinin 2 Receptor (B2R) determined by MAS SSNMR | | Descriptor: | Bradykinin (BK) | | Authors: | Mao, J, Lopez, J.J, Shukla, A.K, Kuenze, G, Meiler, J, Schwalbe, H, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F3W
 
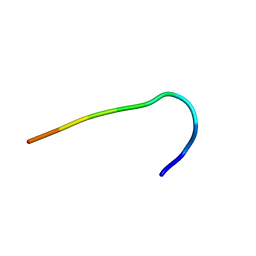 | | Backbone structure of free bradykinin (BK) in DDM/CHS detergent micelle determined by MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Lopez, J.J, Shukla, A.K, Kuenze, G, Meiler, J, Schwalbe, H, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6TKO
 
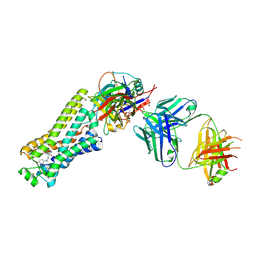 | |
6IBL
 
 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND AGONIST FORMOTEROL AND NANOBODY Nb80 | | Descriptor: | Camelid antibody fragment Nb80, HEGA-10, SODIUM ION, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of beta-arrestin coupling to formoterol-bound beta1-adrenoceptor.
Nature, 583, 2020
|
|
6K3F
 
 | |
8AS3
 
 | | Structure of arrestin2 in complex with 6P CCR5 phosphopeptide and Fab30 | | Descriptor: | Beta-arrestin-1, C-C chemokine receptor type 5, Fab30 heavy chain, ... | | Authors: | Isaikina, P, Jakob, R.P, Maier, T, Grzesiek, S. | | Deposit date: | 2022-08-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A key GPCR phosphorylation motif discovered in arrestin2⋅CCR5 phosphopeptide complexes.
Mol.Cell, 83, 2023
|
|
8AS2
 
 | | Structure of arrestin2 in complex with 4P CCR5 phosphopeptide and Fab30 | | Descriptor: | Beta-arrestin-1, C-C chemokine receptor type 5, Fab30 heavy chain, ... | | Authors: | Isaikina, P, Jakob, R.P, Maier, T, Grzesiek, S. | | Deposit date: | 2022-08-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A key GPCR phosphorylation motif discovered in arrestin2⋅CCR5 phosphopeptide complexes.
Mol.Cell, 83, 2023
|
|
8AS4
 
 | |
