7PYV
 
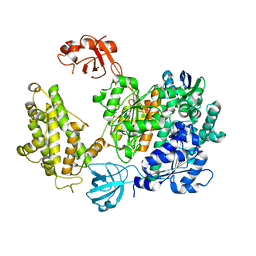 | | Crystal structure of human UBA6 in complex with the ubiquitin-like modifier FAT10 | | Descriptor: | UBD, Ubiquitin-like modifier-activating enzyme 6,Ubiquitin-like modifier-activating enzyme 1,Ubiquitin-like modifier-activating enzyme 6 | | Authors: | Li, S, Truongvan, N, Schindelin, H. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
6ENT
 
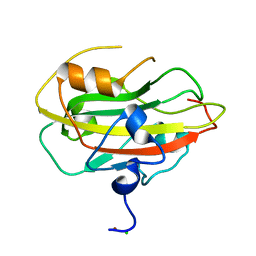 | | Structure of the rat RKIP variant delta143-146 | | Descriptor: | CHLORIDE ION, Phosphatidylethanolamine-binding protein 1, ZINC ION | | Authors: | Koelmel, W, Hirschbeck, M, Schindelin, H, Lorenz, K, Kisker, C. | | Deposit date: | 2017-10-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6ENS
 
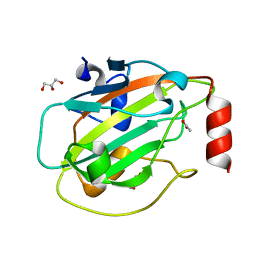 | | Structure of mouse wild-type RKIP | | Descriptor: | ACETATE ION, GLYCEROL, Phosphatidylethanolamine-binding protein 1 | | Authors: | Hirschbeck, M, Koelmel, W, Schindelin, H, Lorenz, K, Kisker, C. | | Deposit date: | 2017-10-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4TK4
 
 | |
4TK2
 
 | |
4TK1
 
 | | Geph E in complex with a GABA receptor alpha3 subunit derived peptide in space group P21212 | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Gamma-aminobutyric acid receptor subunit alpha-3, ... | | Authors: | Kasaragod, V.B, Maric, H.M, Schindelin, H. | | Deposit date: | 2014-05-25 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of the alternative recruitment of GABAA versus glycine receptors through gephyrin.
Nat Commun, 5, 2014
|
|
4TK3
 
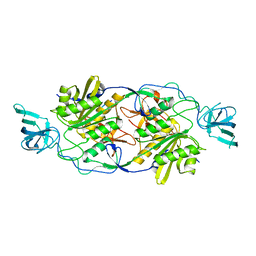 | |
6ZQH
 
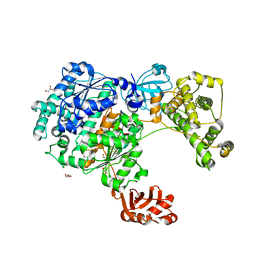 | | Yeast Uba1 in complex with ubiquitin | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | Development of ADPribosyl Ubiquitin Analogues to Study Enzymes Involved in Legionella Infection.
Chemistry, 27, 2021
|
|
4U90
 
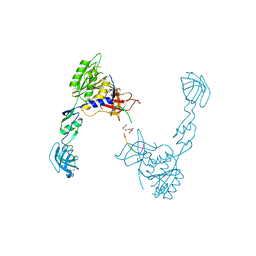 | | GephE in complex with PEG crosslinked GABA receptor alpha3 subunit derived dimeric peptide | | Descriptor: | 1,1'-[ethane-1,2-diylbis(oxyethane-2,1-diyl)]bis(1H-pyrrole-2,5-dione), 1,4-BUTANEDIOL, Gamma-aminobutyric acid receptor subunit alpha-3, ... | | Authors: | Kasaragod, V.B, Maric, H.M, Schindelin, H. | | Deposit date: | 2014-08-05 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of High-Affinity Dimeric Inhibitors Targeting the Interactions between Gephyrin and Inhibitory Neurotransmitter Receptors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4U91
 
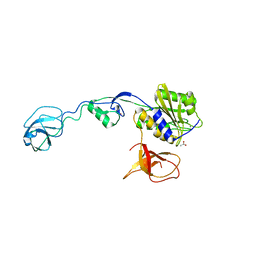 | | GephE in complex with Para-Phenyl crosslinked Glycine receptor beta subunit derived dimeric peptide | | Descriptor: | 1,1'-benzene-1,4-diylbis(1H-pyrrole-2,5-dione), 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Kasaragod, V.B, Maric, H.M, Schindelin, H. | | Deposit date: | 2014-08-05 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of High-Affinity Dimeric Inhibitors Targeting the Interactions between Gephyrin and Inhibitory Neurotransmitter Receptors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7PO7
 
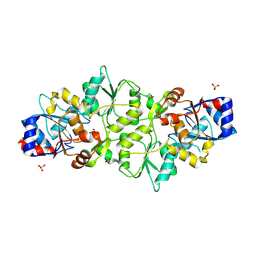 | |
3QQ7
 
 | |
3QQ8
 
 | |
3R3M
 
 | |
3BOA
 
 | |
3IPO
 
 | | Crystal structure of YnjE | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
3IPP
 
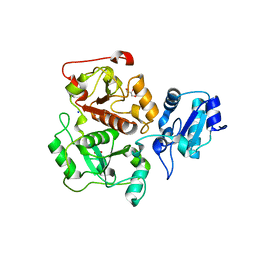 | | crystal structure of sulfur-free YnjE | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative thiosulfate sulfurtransferase ynjE, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
3CMM
 
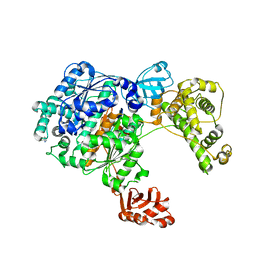 | | Crystal Structure of the Uba1-Ubiquitin Complex | | Descriptor: | PROLINE, Ubiquitin, Ubiquitin-activating enzyme E1 1 | | Authors: | Lee, I, Schindelin, H. | | Deposit date: | 2008-03-23 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes.
Cell(Cambridge,Mass.), 134, 2008
|
|
3ESW
 
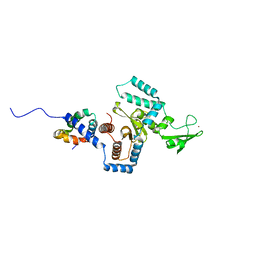 | | Complex of yeast PNGase with GlcNAc2-IAc. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase, UV excision repair protein RAD23, ... | | Authors: | Zhao, G, Zhou, X, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2008-10-06 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and mutational studies on the importance of oligosaccharide binding for the activity of yeast PNGase.
Glycobiology, 19, 2009
|
|
3M63
 
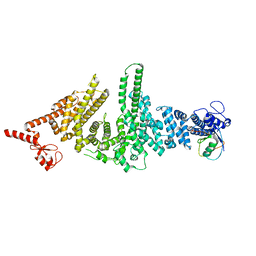 | |
3M62
 
 | |
3GAE
 
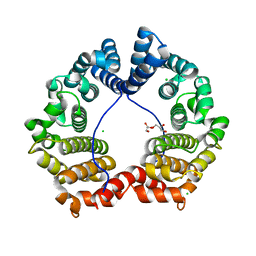 | | Crystal Structure of PUL | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein DOA1 | | Authors: | Zhao, G, Schindelin, H, Lennarz, W.J. | | Deposit date: | 2009-02-17 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Armadillo motif in Ufd3 interacts with Cdc48 and is involved in ubiquitin homeostasis and protein degradation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6YK0
 
 | |
6YJZ
 
 | |
6YK1
 
 | |
