4HZF
 
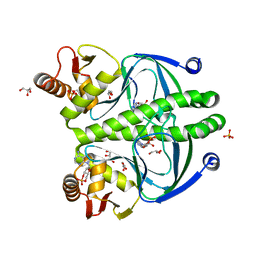 | | structure of the wild type Catabolite gene Activator Protein | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, GLYCEROL, ... | | Authors: | Pohl, E, Townsend, P.D, Rodgers, T, Burnell, D, McLeish, T.C.B, Wilson, M.R, Cann, M. | | Deposit date: | 2012-11-15 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Modulation of global low-frequency motions underlies allosteric regulation: demonstration in CRP/FNR family transcription factors.
Plos Biol., 11, 2013
|
|
4I02
 
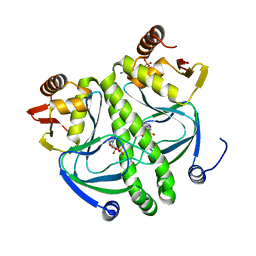 | | structure of the mutant Catabolite gene activator protein V140A | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator | | Authors: | Pohl, E, Townsend, P.D, Rodgers, T, Burnell, D, McLeish, T.C.B, Wilson, M.R, Cann, M.J. | | Deposit date: | 2012-11-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modulation of global low-frequency motions underlies allosteric regulation: demonstration in CRP/FNR family transcription factors.
Plos Biol., 11, 2013
|
|
4I0B
 
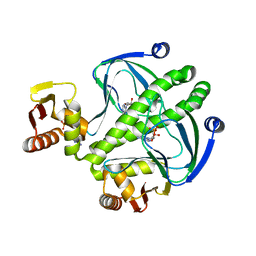 | | structure of the mutant Catabolite gene activator protein H160L | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator | | Authors: | Pohl, E, Townsend, P.D, Rodgers, T, Burnell, D, McLeish, T.C.B, Wilson, M.R, Cann, M.J. | | Deposit date: | 2012-11-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Modulation of global low-frequency motions underlies allosteric regulation: demonstration in CRP/FNR family transcription factors.
Plos Biol., 11, 2013
|
|
7OBO
 
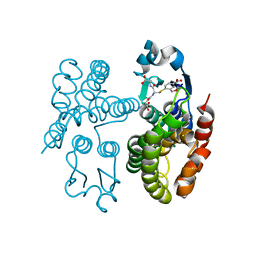 | | GSTF1 from Alopecurus myosuroides | | Descriptor: | (2~{S})-2-azanyl-5-[[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-3-[(7-nitro-2,1,3-benzoxadiazol-4-yl)sulfanyl]-1-oxidanylidene-propan-2-yl]amino]-5-oxidanylidene-pentanoic acid, Glutathione transferase | | Authors: | Pohl, E, Eno, R.F.M, Freitag-Pohl, S. | | Deposit date: | 2021-04-23 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Flavonoid-based inhibitors of the Phi-class glutathione transferase from black-grass to combat multiple herbicide resistance.
Org.Biomol.Chem., 19, 2021
|
|
5NIO
 
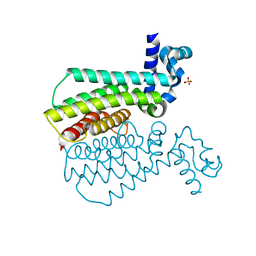 | | EthR complex | | Descriptor: | HTH-type transcriptional regulator EthR, SULFATE ION, ~{N}-butyl-4-methyl-piperidine-1-carboxamide | | Authors: | Pohl, E, Tatum, N.J, Cole, J.C, Baulard, A.R. | | Deposit date: | 2017-03-24 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | New active leads for tuberculosis booster drugs by structure-based drug discovery.
Org. Biomol. Chem., 15, 2017
|
|
5NIZ
 
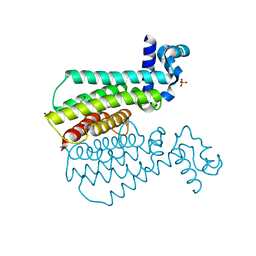 | | EthR complex | | Descriptor: | 1-[3-[5-[(1~{R},2~{R})-2-methylcyclopropyl]furan-2-yl]propanoyl]piperidine-4-carboxamide, HTH-type transcriptional regulator EthR, SULFATE ION | | Authors: | Pohl, E, Tatum, N.J, Cole, J.C, Baulard, A.R. | | Deposit date: | 2017-03-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New active leads for tuberculosis booster drugs by structure-based drug discovery.
Org. Biomol. Chem., 15, 2017
|
|
5NIM
 
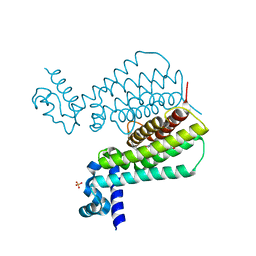 | | EthR complex | | Descriptor: | HTH-type transcriptional regulator EthR, SULFATE ION, [1-(2-hydroxyethyl)pyrrolo[3,4-c]pyrazol-5-yl]-(5-propyl-1,2-oxazol-3-yl)methanone | | Authors: | Pohl, E, Tatum, N.J, Cole, J.C, Baulard, A.R. | | Deposit date: | 2017-03-24 | | Release date: | 2017-11-15 | | Last modified: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New active leads for tuberculosis booster drugs by structure-based drug discovery.
Org. Biomol. Chem., 15, 2017
|
|
5NJ0
 
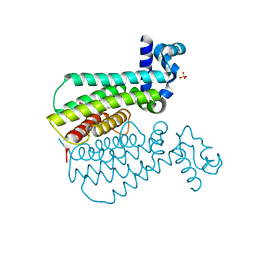 | | EthR complex | | Descriptor: | HTH-type transcriptional regulator EthR, SULFATE ION, ~{N}-[(2~{R})-2-(4-nitrophenyl)-2-oxidanyl-ethyl]-1,3-benzodioxole-5-carboxamide | | Authors: | Pohl, E, Tatum, N.J, Cole, J.C, Baulard, A.R. | | Deposit date: | 2017-03-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New active leads for tuberculosis booster drugs by structure-based drug discovery.
Org. Biomol. Chem., 15, 2017
|
|
7ODM
 
 | | AmGSTF1 Y118S variant | | Descriptor: | Glutathione transferase, [(2~{S})-5-[[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-3-sulfanyl-propan-2-yl]amino]-1-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]azanium | | Authors: | Pohl, E, Eno, R.F.M. | | Deposit date: | 2021-04-30 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Flavonoid-based inhibitors of the Phi-class glutathione transferase from black-grass to combat multiple herbicide resistance.
Org.Biomol.Chem., 19, 2021
|
|
4BYY
 
 | | Apo GlxR | | Descriptor: | GLYCEROL, PHOSPHATE ION, TRANSCRIPTIONAL REGULATOR, ... | | Authors: | Pohl, E, Cann, M.J, Townsend, P.D, Money, V.A. | | Deposit date: | 2013-07-21 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The Crystal Structures of Apo and Camp-Bound Glxr from Corynebacterium Glutamicum Reveal Structural and Dynamic Changes Upon Camp Binding in Crp/Fnr Family Transcription Factors.
Plos One, 9, 2014
|
|
4CMY
 
 | | Chlorobium tepidum Ferritin | | Descriptor: | FE (III) ION, FERRITIN | | Authors: | Pohl, E, Arenas, M, Townsend, P.D, Yevenes, A. | | Deposit date: | 2014-01-18 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The Crystal Structure of Ferritin from Chlorobium Tepidum Reveals a New Conformation of the 4-Fold Channel for This Protein Family.
Biochimie, 106, 2014
|
|
7Z3M
 
 | |
6IA5
 
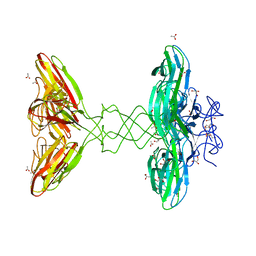 | | Crystal Structure Analysis of Bacillus subtilis 168 XepA | | Descriptor: | ACETATE ION, GLYCEROL, Phage-like element PBSX protein XepA | | Authors: | Freitag-Pohl, S, Pohl, E. | | Deposit date: | 2018-11-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of the Bacillus subtilis prophage lytic cassette proteins XepA and YomS.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8B9W
 
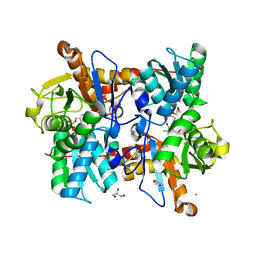 | | Cysteine Synthase from Trypanosoma theileri with PLP bound | | Descriptor: | CHLORIDE ION, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Sowerby, K.V, Pohl, E. | | Deposit date: | 2022-10-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Cysteine synthase: multiple structures of a key enzyme in cysteine synthesis and a potential drug target for Chagas disease and leishmaniasis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B9Y
 
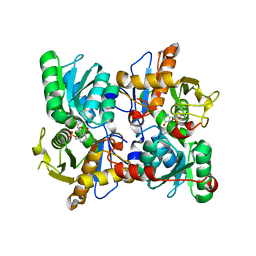 | | Cysteine Synthase from Trypanosoma cruzi with PLP and OAS | | Descriptor: | Cysteine synthase, putative, GLYCEROL, ... | | Authors: | Sowerby, K.V, Pohl, E. | | Deposit date: | 2022-10-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cysteine synthase: multiple structures of a key enzyme in cysteine synthesis and a potential drug target for Chagas disease and leishmaniasis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B9M
 
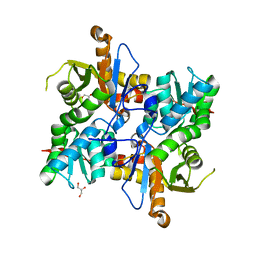 | | Cysteine Synthase from Leishmania Infantum | | Descriptor: | CHLORIDE ION, Cysteine synthase, GLYCEROL | | Authors: | Sowerby, K.V, Pohl, E. | | Deposit date: | 2022-10-06 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cysteine synthase: multiple structures of a key enzyme in cysteine synthesis and a potential drug target for Chagas disease and leishmaniasis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
2YCE
 
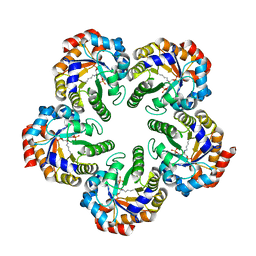 | | Structure of an Archaeal fructose-1,6-bisphosphate aldolase with the catalytic Lys covalently bound to the carbinolamine intermediate of the substrate. | | Descriptor: | D-MANNITOL-1,6-DIPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS 1 | | Authors: | Lorentzen, E, Siebers, B, Hensel, R, Pohl, E. | | Deposit date: | 2011-03-14 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanism of the Schiff Base Forming Fructose-1,6-Bisphosphate Aldolase: Structural Analysis of Reaction Intermediates.
Biochemistry, 44, 2005
|
|
7B3N
 
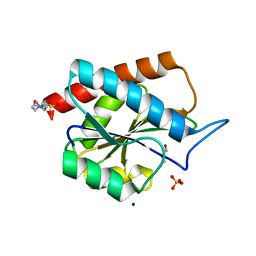 | | AmiP amidase-3 from Thermus parvatiensis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Cell wall hydrolase, ... | | Authors: | Freitag-Pohl, S, Pohl, E. | | Deposit date: | 2020-12-01 | | Release date: | 2022-06-22 | | Last modified: | 2023-05-10 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | AmiP from hyperthermophilic Thermus parvatiensis prophage is a thermoactive and ultrathermostable peptidoglycan lytic amidase.
Protein Sci., 32, 2023
|
|
7PJO
 
 | |
5FMN
 
 | |
3MBU
 
 | | Structure of a bipyridine-modified PNA duplex | | Descriptor: | 1,2-ETHANEDIOL, Bipyridine-PNA, CARBONATE ION, ... | | Authors: | Yeh, J.I, Pohl, E, Truan, D, He, W, Sheldrick, G.M, Du, S, Achim, C. | | Deposit date: | 2010-03-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The crystal structure of non-modified and bipyridine-modified PNA duplexes.
Chemistry, 16, 2010
|
|
3MBS
 
 | | Crystal structure of 8mer PNA | | Descriptor: | 1,2-ETHANEDIOL, Peptide Nucleic Acid | | Authors: | Yeh, J.I, Pohl, E, Truan, D, He, W, Sheldrick, G.M, Achim, C. | | Deposit date: | 2010-03-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | The crystal structure of non-modified and bipyridine-modified PNA duplexes.
Chemistry, 16, 2010
|
|
6HKR
 
 | | Human Cellular Retinoic Acid Binding Protein II (CRABPII) with bound synthetic retinoid DC271. | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4,4-dimethyl-1-propan-2-yl-2,3-dihydroquinolin-6-yl)ethynyl]benzoic acid, Cellular retinoic acid-binding protein 2, ... | | Authors: | Tomlinson, C, Chisholm, D, Whiting, A, Pohl, E. | | Deposit date: | 2018-09-07 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel Fluorescence Competition Assay for Retinoic Acid Binding Proteins.
ACS Med Chem Lett, 9, 2018
|
|
4WGF
 
 | | YcaC from Pseudomonas aeruginosa with hexane-2,5-diol and covalent acrylamide | | Descriptor: | (2R,5R)-hexane-2,5-diol, CHLORIDE ION, PROPIONAMIDE, ... | | Authors: | Groftehauge, M.K, Truan, D, Vasil, A, Denny, P.W, Vasil, M.L, Pohl, E. | | Deposit date: | 2014-09-18 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34020686 Å) | | Cite: | Crystal Structure of a Hidden Protein, YcaC, a Putative Cysteine Hydrolase from Pseudomonas aeruginosa, with and without an Acrylamide Adduct.
Int J Mol Sci, 16, 2015
|
|
4WH0
 
 | | YcaC from Pseudomonas aeruginosa with S-mercaptocysteine active site cysteine | | Descriptor: | CHLORIDE ION, Putative hydrolase | | Authors: | Groftehauge, M.K, Truan, D, Vasil, A, Denny, P.W, Vasil, M.L, Pohl, E. | | Deposit date: | 2014-09-19 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.563 Å) | | Cite: | Crystal Structure of a Hidden Protein, YcaC, a Putative Cysteine Hydrolase from Pseudomonas aeruginosa, with and without an Acrylamide Adduct.
Int J Mol Sci, 16, 2015
|
|
