8A8T
 
 | |
8A7S
 
 | |
8A30
 
 | |
8AEQ
 
 | |
8AEO
 
 | |
8AET
 
 | |
8AEW
 
 | |
8AER
 
 | |
5M45
 
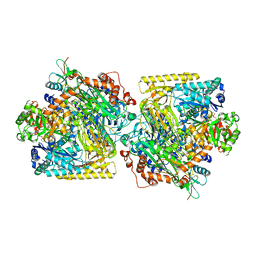 | | Structure of Acetone Carboxylase purified from Xanthobacter autotrophicus | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ACETATE ION, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kabasakal, B.V, Wells, J.N, Nwaobi, B.C, Eilers, B.J, Peters, J.W, Murray, J.W. | | Deposit date: | 2016-10-18 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
6HGB
 
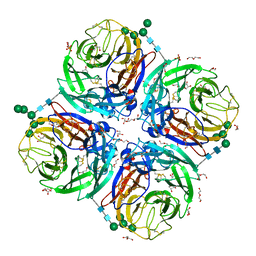 | | Influenza A virus N6 neuraminidase native structure (Duck/England/56). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-23 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
6HG0
 
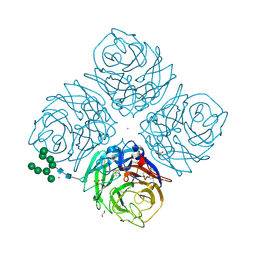 | | Influenza A Virus N9 Neuraminidase complex with NANA (Tern/Australia). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
6HEB
 
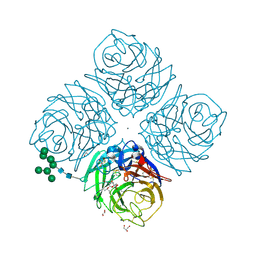 | | Influenza A Virus N9 Neuraminidase complex with Oseltamivir (Tern). | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-20 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site.
To Be Published
|
|
6HFC
 
 | | Influenza A Virus N9 Neuraminidase Native (Tern). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
6HCX
 
 | | Influenza Virus N9 Neuraminidase A complex with Zanamivir molecule (Tern). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
6HFY
 
 | | Influenza A virus N6 neuraminidase complex with DANA (Duck/England/56). | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
6HG5
 
 | | Influenza A virus N6 neuraminidase complex with Oseltamivir (Duck/England/56). | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-22 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site.
To Be Published
|
|
6HUF
 
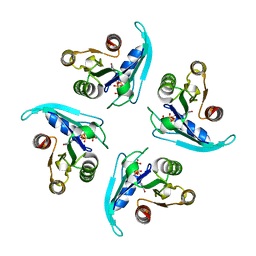 | | Coping with strong translational non-crystallographic symmetry and extreme anisotropy in molecular replacement with Phaser: human Rab27a | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-27A | | Authors: | Jamshidiha, M, Perez-Dorado, I, Murray, J.W, Tate, E.W, Cota, E, Read, R.J. | | Deposit date: | 2018-10-08 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Coping with strong translational noncrystallographic symmetry and extreme anisotropy in molecular replacement with Phaser: human Rab27a.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8AM5
 
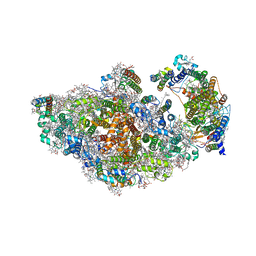 | | RCII/PSI complex, class 3 | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, Z, Vercellino, I, Knoppova, J, Sobotka, R, Murray, J.W, Nixon, P.J, Sazanov, L.A, Komenda, J. | | Deposit date: | 2022-08-02 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Ycf48 accessory factor occupies the site of the oxygen-evolving manganese cluster during photosystem II biogenesis.
Nat Commun, 14, 2023
|
|
8ASL
 
 | | RCII/PSI complex, class 2 | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, Z, Vercellino, I, Knoppova, J, Sobotka, R, Murray, J.W, Nixon, P.J, Sazanov, L.A, Komenda, J. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | The Ycf48 accessory factor occupies the site of the oxygen-evolving manganese cluster during photosystem II biogenesis.
Nat Commun, 14, 2023
|
|
8ASP
 
 | | RCII/PSI complex, focused refinement of PSI | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, Z, Vercellino, I, Knoppova, J, Sobotka, R, Murray, J.W, Nixon, P.J, Sazanov, L.A, Komenda, J. | | Deposit date: | 2022-08-20 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The Ycf48 accessory factor occupies the site of the oxygen-evolving manganese cluster during photosystem II biogenesis.
Nat Commun, 14, 2023
|
|
5LYP
 
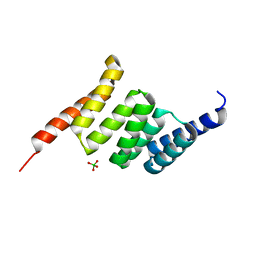 | | Crystal structure of the Tpr Domain of Sgt2. | | Descriptor: | BORATE ION, Small glutamine-rich tetratricopeptide repeat-containing protein 2 | | Authors: | Krysztofinska, E.M, Morgan, R.M.L, Thapaliya, A, Evans, N.J, Murray, J.W, Martinez-Lumbreras, S, Isaacson, R.L. | | Deposit date: | 2016-09-28 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and Interactions of the TPR Domain of Sgt2 with Yeast Chaperones and Ybr137wp.
Front Mol Biosci, 4, 2017
|
|
6SDM
 
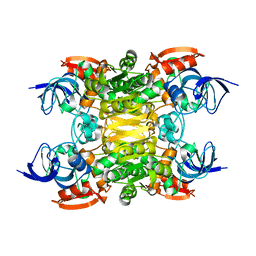 | | NADH-dependent variant of TBADH | | Descriptor: | NADP-dependent isopropanol dehydrogenase, ZINC ION | | Authors: | Selles Vidal, L, Murray, J.W, Heap, J.T. | | Deposit date: | 2019-07-28 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Versatile selective evolutionary pressure using synthetic defect in universal metabolism.
Nat Commun, 12, 2021
|
|
6SCH
 
 | | NADH-dependent variant of CBADH | | Descriptor: | MAGNESIUM ION, NADP-dependent isopropanol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Selles Vidal, L, Murray, J.W, Heap, J.T. | | Deposit date: | 2019-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Versatile selective evolutionary pressure using synthetic defect in universal metabolism.
Nat Commun, 12, 2021
|
|
6TRG
 
 | | Salmonella typhimurium neuraminidase mutant (D100S) | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sialidase | | Authors: | Garman, E.F, Salinger, M.T, Murray, J.W, Laver, W.G, Kuhn, P, Vimr, E.R. | | Deposit date: | 2019-12-18 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Salmonella typhimurium neuraminidase mutant (D100S)
To Be Published
|
|
6TVI
 
 | | Salmonella typhimurium mutant neuraminidase (D100S)+ DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, Sialidase | | Authors: | Garman, E.F, Salinger, M.T, Murray, J.W, Laver, W.G, Kuhn, P, Vimr, E.R. | | Deposit date: | 2020-01-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Salmonella typhimurium mutant neuraminidase (D100S)+ DANA
To Be Published
|
|
