4PD9
 
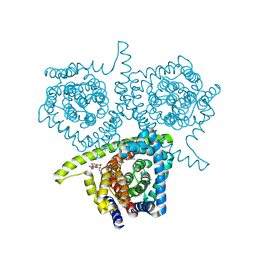 | | Structure of vcCNT-7C8C bound to adenosine | | Descriptor: | ADENOSINE, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | Authors: | Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2014-04-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.096 Å) | | Cite: | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
6BW5
 
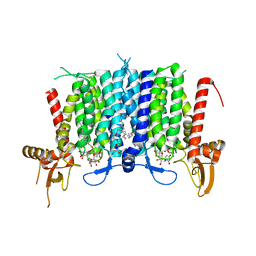 | | Human GPT (DPAGT1) in complex with tunicamycin | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Tunicamycin, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Yoo, J, Kuk, A.C.Y, Mashalidis, E.H, Lee, S.-Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | GlcNAc-1-P-transferase-tunicamycin complex structure reveals basis for inhibition of N-glycosylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6BPQ
 
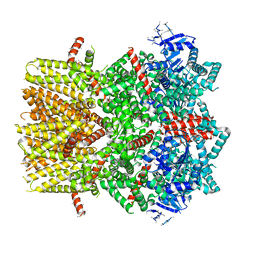 | | Structure of the cold- and menthol-sensing ion channel TRPM8 | | Descriptor: | Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Wu, M, Zubcevic, L, Borschel, W.F, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2017-11-25 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the cold- and menthol-sensing ion channel TRPM8.
Science, 359, 2018
|
|
6BW6
 
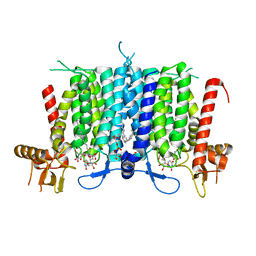 | | Human GPT (DPAGT1) H129 variant in complex with tunicamycin | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Tunicamycin, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Yoo, J, Kuk, A.C.Y, Mashalidis, E.H, Lee, S.-Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | GlcNAc-1-P-transferase-tunicamycin complex structure reveals basis for inhibition of N-glycosylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DT0
 
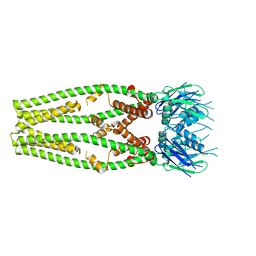 | | Cryo-EM structure of a mitochondrial calcium uniporter | | Descriptor: | CALCIUM ION, Mitochondrial calcium uniporter | | Authors: | Yoo, J, Wu, M, Yin, Y, Herzik, M.A.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2018-06-14 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a mitochondrial calcium uniporter.
Science, 361, 2018
|
|
3TIJ
 
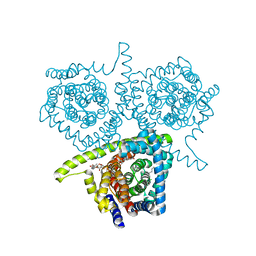 | | Crystal structure of a concentrative nucleoside transporter from Vibrio cholerae | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, SODIUM ION, ... | | Authors: | Johnson, Z.L, Cheong, C.-G, Lee, S.-Y. | | Deposit date: | 2011-08-20 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Crystal structure of a concentrative nucleoside transporter from Vibrio cholerae at 2.4A
Nature, 483, 2012
|
|
5HZ2
 
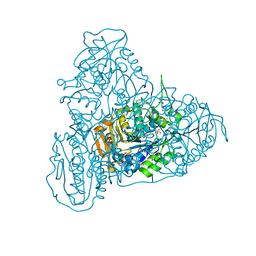 | | Crystal structure of PhaC1 from Ralstonia eutropha | | Descriptor: | GLYCEROL, Poly-beta-hydroxybutyrate polymerase, SULFATE ION | | Authors: | Kim, J, Kim, K.-J. | | Deposit date: | 2016-02-02 | | Release date: | 2016-12-07 | | Last modified: | 2017-04-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms.
Biotechnol J, 12, 2017
|
|
4C1D
 
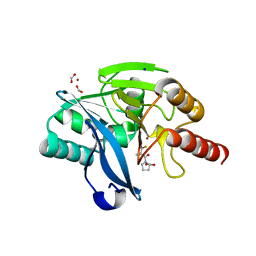 | | Crystal structure of the metallo-beta-lactamase VIM-2 with L-captopril | | Descriptor: | BETA-LACTAMASE CLASS B VIM-2, FORMIC ACID, L-CAPTOPRIL, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
4C1C
 
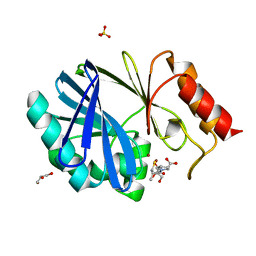 | | Crystal structure of the metallo-beta-lactamase BCII with D-captopril | | Descriptor: | 1-(3-MERCAPTO-2-METHYL-PROPIONYL)-PYRROLIDINE-2-CARBOXYLIC ACID, BETA-LACTAMASE 2, GLYCEROL, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
5U9W
 
 | | Structure of CNTnw N149L in the intermediate 3 state | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-2-octyldecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Nucleoside permease | | Authors: | Hirschi, M, Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2016-12-18 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.555 Å) | | Cite: | Visualizing multistep elevator-like transitions of a nucleoside transporter.
Nature, 545, 2017
|
|
4C1E
 
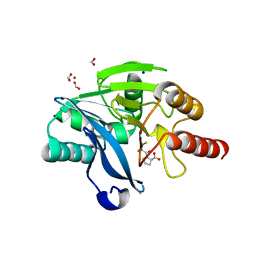 | | Crystal structure of the metallo-beta-lactamase VIM-2 with D-captopril | | Descriptor: | 1-(3-MERCAPTO-2-METHYL-PROPIONYL)-PYRROLIDINE-2-CARBOXYLIC ACID, BETA-LACTAMASE CLASS B VIM-2, FORMIC ACID, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
4C09
 
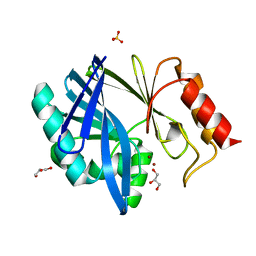 | | Crystal structure of the metallo-beta-lactamase BCII | | Descriptor: | BETA-LACTAMASE 2, GLYCEROL, SULFATE ION, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
4C1G
 
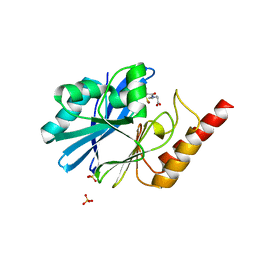 | | Crystal structure of the metallo-beta-lactamase IMP-1 with D-captopril | | Descriptor: | 1-(3-MERCAPTO-2-METHYL-PROPIONYL)-PYRROLIDINE-2-CARBOXYLIC ACID, BETA-LACTAMASE IMP-1, SULFATE ION, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.714 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
4BZ3
 
 | | Crystal structure of the metallo-beta-lactamase VIM-2 | | Descriptor: | BETA-LACTAMASE VIM-2, FORMIC ACID, SODIUM ION, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-07-23 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.294 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
4C1F
 
 | | Crystal structure of the metallo-beta-lactamase IMP-1 with L-captopril | | Descriptor: | BETA-LACTAMASE IMP-1, L-CAPTOPRIL, SULFATE ION, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
4C1H
 
 | | Crystal structure of the metallo-beta-lactamase BCII with L-captopril | | Descriptor: | BETA-LACTAMASE 2, GLYCEROL, L-CAPTOPRIL, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
3AY3
 
 | |
5K48
 
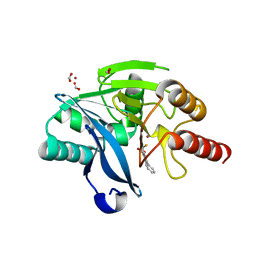 | | VIM-2 Metallo Beta Lactamase in complex with 3-(mercaptomethyl)-[1,1'-biphenyl]-4-carboxylic acid | | Descriptor: | 4-phenyl-2-(sulfanylmethyl)benzoic acid, Beta-lactamase VIM-2, FORMIC ACID, ... | | Authors: | Zollman, D, McDonough, M, Brem, J, Schofield, C. | | Deposit date: | 2016-05-20 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | In Silico Fragment-Based Design Identifies Subfamily B1 Metallo-beta-lactamase Inhibitors.
J. Med. Chem., 61, 2018
|
|
6NR4
 
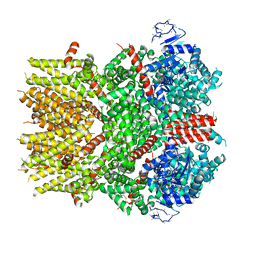 | | Cryo-EM structure of the TRPM8 ion channel with low occupancy icilin, PI(4,5)P2, and calcium | | Descriptor: | Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | Deposit date: | 2019-01-22 | | Release date: | 2019-02-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
6NR2
 
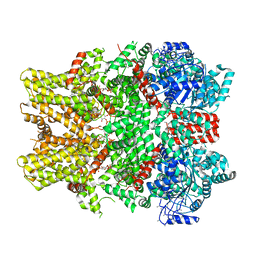 | | Cryo-EM structure of the TRPM8 ion channel in complex with the menthol analog WS-12 and PI(4,5)P2 | | Descriptor: | (1R,2S,5R)-N-(4-methoxyphenyl)-5-methyl-2-(propan-2-yl)cyclohexane-1-carboxamide, (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl icosa-5,8,11,14-tetraenoate, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | Deposit date: | 2019-01-22 | | Release date: | 2019-02-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
6NR3
 
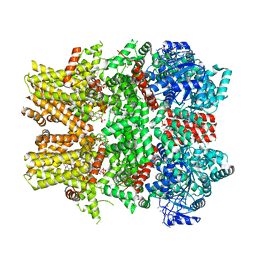 | | Cryo-EM structure of the TRPM8 ion channel in complex with high occupancy icilin, PI(4,5)P2, and calcium | | Descriptor: | (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl icosa-5,8,11,14-tetraenoate, CALCIUM ION, Icilin, ... | | Authors: | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | Deposit date: | 2019-01-22 | | Release date: | 2019-02-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
6NC7
 
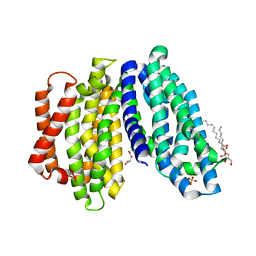 | | Lipid II flippase MurJ, inward open conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lipid II flippase MurJ, SULFATE ION | | Authors: | Kuk, A.C.Y, Lee, S.-Y. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Visualizing conformation transitions of the Lipid II flippase MurJ.
Nat Commun, 10, 2019
|
|
6NC6
 
 | | Lipid II flippase MurJ, inward closed conformation | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Lipid II flippase MurJ, ... | | Authors: | Kuk, A.C.Y, Lee, S.-Y. | | Deposit date: | 2018-12-10 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Visualizing conformation transitions of the Lipid II flippase MurJ.
Nat Commun, 10, 2019
|
|
6NC8
 
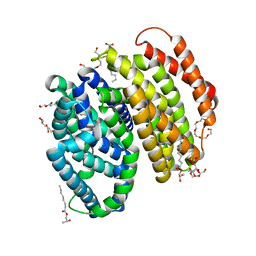 | | Lipid II flippase MurJ, inward occluded conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lipid II flippase MurJ, PENTAETHYLENE GLYCOL, ... | | Authors: | Kuk, A.C.Y, Lee, S.-Y. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Visualizing conformation transitions of the Lipid II flippase MurJ.
Nat Commun, 10, 2019
|
|
6NC9
 
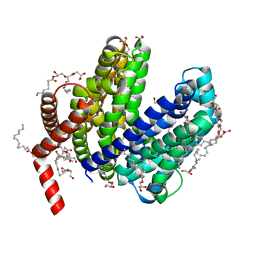 | | Lipid II flippase MurJ, outward-facing conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lipid II flippase MurJ, ... | | Authors: | Kuk, A.C.Y, Lee, S.-Y. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing conformation transitions of the Lipid II flippase MurJ.
Nat Commun, 10, 2019
|
|
