5ZFJ
 
 | | Crystal structure of a cyclase Filc from Fischerella sp. in complex with 4-(1H-Indol-3-yl)butan-2-one | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(1~{H}-indol-3-yl)butan-2-one, CALCIUM ION, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-03-06 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6A98
 
 | | Crystal structure of a cyclase from Fischerella sp. TAU | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, aromatic prenyltransferase, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-12 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5ZZD
 
 | | Crystal structure of a protein from Aspergillus flavus | | Descriptor: | O-methyltransferase lepI, S-ADENOSYLMETHIONINE | | Authors: | Chang, Z.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-05-31 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of LepI, a multifunctional SAM-dependent enzyme which catalyzes pericyclic reactions in leporin biosynthesis.
Org.Biomol.Chem., 17, 2019
|
|
6A8X
 
 | | Crystal structure of a apo form cyclase from Fischerella sp. | | Descriptor: | CALCIUM ION, aromatic prenyltransferase | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-11 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6A92
 
 | | Crystal structure of a cyclase Filc1 from Fischerella sp. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-11 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5ZXA
 
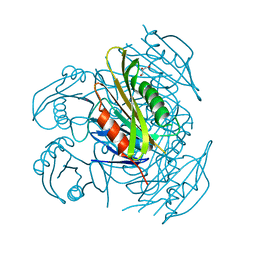 | | Crystal structure of fibronectin-binding protein Apa mutant from Mycobacterium tuberculosis | | Descriptor: | Alanine and proline-rich secreted protein Apa, GLYCEROL, MERCURY (II) ION | | Authors: | Gao, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-05-18 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Functional and structural investigations of fibronectin-binding protein Apa from Mycobacterium tuberculosis.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
6AK9
 
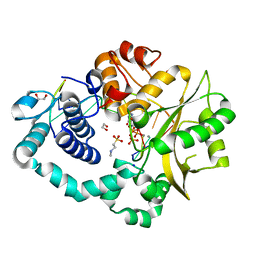 | | Pre-catalytic Ternary Complex of Human DNA Polymerase Mu with Templating Cytosine and Incoming Ca-8oxodGTP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-08-30 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
5ZX9
 
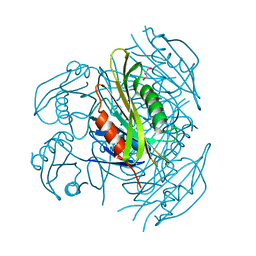 | | Crystal structure of apo form fibronectin-binding protein Apa from Mycobacterium tuberculosis | | Descriptor: | Alanine and proline-rich secreted protein Apa, GLYCEROL | | Authors: | Gao, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-05-18 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Functional and structural investigations of fibronectin-binding protein Apa from Mycobacterium tuberculosis.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
6AK5
 
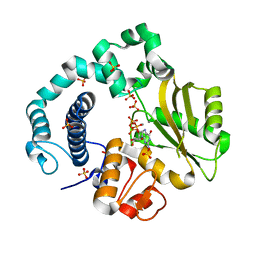 | | Binary Complex of Human DNA Polymerase Mu with MnGTP | | Descriptor: | DNA-directed DNA/RNA polymerase mu, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-08-30 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6AEC
 
 | | Binary complex of human DNA Polymerase Mu with MnATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-directed DNA/RNA polymerase mu, MANGANESE (II) ION, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-08-04 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6AK8
 
 | | Pre-catalytic Ternary Complex of Human DNA Polymerase Mu with Templating Adenine and Incoming Ca-8oxodGTP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-08-30 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6A99
 
 | | Crystal structure of a Stig cyclases Fisc from Fischerella sp. TAU in complex with (3Z)-3-(1-methyl-2-pyrrolidinylidene)-3H-indole | | Descriptor: | (3~{Z})-3-(1-methylpyrrolidin-2-ylidene)indole, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-12 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6ADU
 
 | | Crystal structure of an enzyme in complex with ligand C | | Descriptor: | (3~{Z})-3-(1-methylpyrrolidin-2-ylidene)indole, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Tan, X.K, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-08-02 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement.
Angew.Chem.Int.Ed.Engl., 57, 2018
|
|
2ZU3
 
 | | Complex structure of CVB3 3C protease with TG-0204998 | | Descriptor: | 3C proteinase, N-[(benzyloxy)carbonyl]-3-[(2,2-dimethylpropanoyl)amino]-L-alanyl-N-[(1R)-4-oxo-1-{[(3S)-2-oxopyrrolidin-3-yl]methyl}pentyl]-L-leucinamide | | Authors: | Lee, C.C, Tsui, Y.C, Wang, A.H.-J. | | Deposit date: | 2008-10-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2ZTX
 
 | | Complex structure of CVB3 3C protease with EPDTC | | Descriptor: | 3C proteinase, zinc(II)hydrogensulfide | | Authors: | Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2ZU5
 
 | | complex structure of SARS-CoV 3CL protease with TG-0205486 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-[(1R)-4-cyclopropyl-4-oxo-1-{[(3S)-2-oxopyrrolidin-3-yl]methyl}butyl]-L-leucinamide | | Authors: | Hsu, M.F, Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2008-10-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2ZU1
 
 | |
3ADZ
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with intermediate PSPP | | Descriptor: | Dehydrosqualene synthase, MAGNESIUM ION, {(1R,2R,3R)-2-[(3E)-4,8-dimethylnona-3,7-dien-1-yl]-2-methyl-3-[(1E,5E)-2,6,10-trimethylundeca-1,5,9-trien-1-yl]cyclopropyl}methyl trihydrogen diphosphate | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AE0
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with geranylgeranyl thiopyrophosphate | | Descriptor: | Dehydrosqualene synthase, MAGNESIUM ION, phosphonooxy-[(10E)-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraenyl]sulfanyl-phosphinic acid | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2ZU2
 
 | | complex structure of CoV 229E 3CL protease with EPDTC | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3C-like proteinase, zinc(II)hydrogensulfide | | Authors: | Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2008-10-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2ZTY
 
 | |
2ZU4
 
 | | Complex structure of SARS-CoV 3CL protease with TG-0204998 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-[(2,2-dimethylpropanoyl)amino]-L-alanyl-N-[(1R)-4-oxo-1-{[(3S)-2-oxopyrrolidin-3-yl]methyl}pentyl]-L-leucinamide | | Authors: | Hsu, M.F, Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2008-10-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
3ACW
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with BPH-651 | | Descriptor: | (3R)-3-biphenyl-4-yl-1-azabicyclo[2.2.2]octan-3-ol, Dehydrosqualene synthase | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-13 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2ZTZ
 
 | |
3ACX
 
 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with BPH-673 | | Descriptor: | Dehydrosqualene synthase, N-(1-methylethyl)-3-[(3-prop-2-en-1-ylbiphenyl-4-yl)oxy]propan-1-amine | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-13 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
