4R4H
 
 | | Crystal structure of non-neutralizing, A32-like antibody 2.2c in complex with HIV-1 Env gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2.2c, Heavy chain, ... | | Authors: | Mclellan, J, Acharya, P, Huang, C.-C, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-09-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.28 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
4R4N
 
 | | Crystal structure of the anti-hiv-1 antibody 2.2c in complex with hiv-1 93ug037 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2.2c LIGHT CHAIN, Antibody 2.2c heavy CHAIN, ... | | Authors: | Acharya, P, Louder, R, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
4R4F
 
 | | Crystal structure of non-neutralizing, A32-like antibody 2.2c in complex with HIV-1 YU2 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2.2c LIGHT CHAIN, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-09-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.514 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
4RQS
 
 | | Crystal structure of fully glycosylated HIV-1 gp120 core bound to CD4 and 17b Fab | | Descriptor: | 17b Fab Heavy Chain, 17b Fab Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kong, L, Wilson, I.A, Kwong, P.D. | | Deposit date: | 2014-11-05 | | Release date: | 2014-12-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.493 Å) | | Cite: | Crystal structure of a fully glycosylated HIV-1 gp120 core reveals a stabilizing role for the glycan at Asn262.
Proteins, 83, 2015
|
|
7Z7C
 
 | |
2RLL
 
 | | CCR5 Nt(7-15) | | Descriptor: | 9-mer from C-C chemokine receptor type 5 | | Authors: | Bewley, C.A, Lam, S.N. | | Deposit date: | 2007-07-21 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4
Science, 317, 2007
|
|
6XEY
 
 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-4 | | Descriptor: | 2-4 Heavy Chain, 2-4 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rapp, M, Shapiro, L, Ho, D.D. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-22 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike.
Nature, 584, 2020
|
|
4NGH
 
 | |
4NHC
 
 | |
7SI2
 
 | |
7SD5
 
 | | Crystallographic structure of neutralizing antibody 10-40 in complex with SARS-CoV-2 spike receptor binding domain | | Descriptor: | 10-40 Heavy chain, 10-40 Light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Reddem, E.R, Casner, R.G, Shapiro, L. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | An antibody class with a common CDRH3 motif broadly neutralizes sarbecoviruses.
Sci Transl Med, 14, 2022
|
|
8AED
 
 | |
6UDA
 
 | | Cryo-EM structure of CH235UCA bound to Man5-enriched CH505.N279K.G458Y.SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH235 UCA heavy chain Fab, CH235 UCA light chain Fab, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2019-09-19 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Neutralization-guided design of HIV-1 envelope trimers with high affinity for the unmutated common ancester of CH235 lineage CD4bs broadly neutralizing antibodies.
Plos Pathog., 15, 2019
|
|
6WRP
 
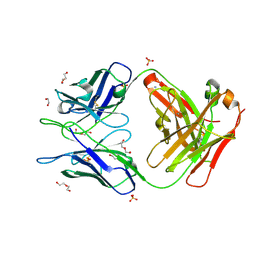 | | Crystal Structure of PI3-E12 Fab, An Antibody Against Human Parainfluenza Virus Type III | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Weidle, C, Pancera, M. | | Deposit date: | 2020-04-29 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Protective antibodies against human parainfluenza virus type 3 infection.
Mabs, 13, 2021
|
|
6X58
 
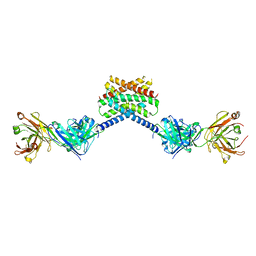 | | MPER-Fluc-Ec2 bound to 10E8v4 antibody | | Descriptor: | 10E8v4 Fab Heavy Chain, 10E8v4 Fab Light Chain, gp41 MPER peptide,Putative fluoride ion transporter CrcB | | Authors: | McIlwain, B.C, Stockbridge, R.B. | | Deposit date: | 2020-05-25 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | N-terminal Transmembrane-Helix Epitope Tag for X-ray Crystallography and Electron Microscopy of Small Membrane Proteins.
J.Mol.Biol., 433, 2021
|
|
6XCJ
 
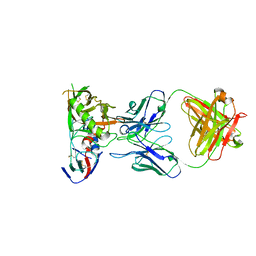 | | Crystal Structure of DH650 Fab from a Rhesus Macaque in Complex with HIV-1 gp120 Core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH650 Fab Heavy Chain, DH650 Fab Light Chain, ... | | Authors: | Raymond, D.D, Chug, H, Harrison, S.C. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recapitulation of HIV-1 Env-antibody coevolution in macaques leading to neutralization breadth.
Science, 371, 2021
|
|
8EUW
 
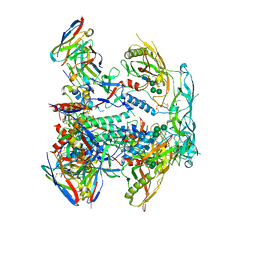 | | Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-MM28 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Pletnev, S, Kwong, P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
8EUU
 
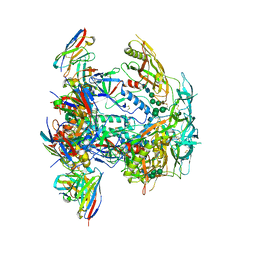 | | Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Pletnev, S, Kwong, P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
6XRJ
 
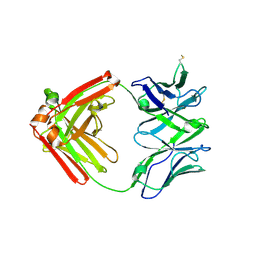 | |
8EUV
 
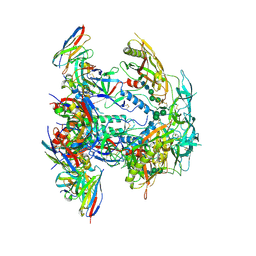 | | Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-COMBO1 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Pletnev, S, Kwong, P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
5JNY
 
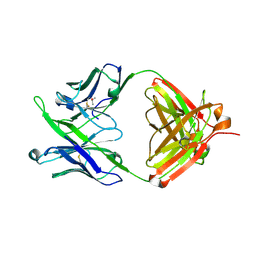 | | Crystal Structure of 10E8 Fab | | Descriptor: | 10E8 Heavy Chain, 10E8 Light Chain, CHLORIDE ION, ... | | Authors: | Ofek, G, Kwong, P. | | Deposit date: | 2016-05-01 | | Release date: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (3.041 Å) | | Cite: | Developmental Pathway of the MPER-Directed HIV-1-Neutralizing Antibody 10E8.
Plos One, 11, 2016
|
|
4GW4
 
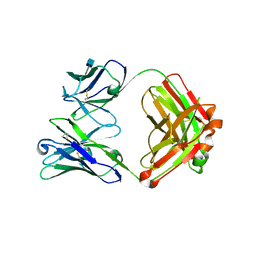 | | Crystal structure of 3BNC60 Fab with P61A mutation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC60 Fab Heavy-chain, 3BNC60 Fab Light-chain | | Authors: | Diskin, R, Fu, B.Z, Bjorkman, P.J. | | Deposit date: | 2012-08-31 | | Release date: | 2013-04-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Somatic Mutations of the Immunoglobulin Framework Are Generally Required for Broad and Potent HIV-1 Neutralization.
Cell(Cambridge,Mass.), 153, 2013
|
|
7RW2
 
 | |
8DEC
 
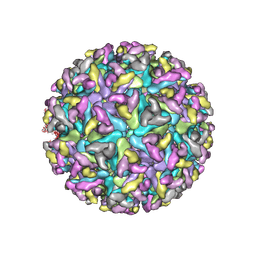 | | Cryo-EM Structure of Western Equine Encephalitis Virus | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2 | | Authors: | Pletnev, S, Verardi, R, Roedeger, M, Kwong, P. | | Deposit date: | 2022-06-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
8DED
 
 | | Cryo-EM Structure of Western Equine Encephalitis Virus VLP in complex with SKW19 fab | | Descriptor: | SKW19 Fab heavy chain, SKW19 Fab light chain, Spike glycoprotein E1, ... | | Authors: | Pletnev, S, Tsybovsky, Y, Verardi, R, Roedeger, M, Kwong, P. | | Deposit date: | 2022-06-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
