1YVX
 
 | | Hepatitis C Virus RNA Polymerase Genotype 2a In Complex With Non- Nucleoside Analogue Inhibitor | | Descriptor: | 3-[ISOPROPYL(4-METHYLBENZOYL)AMINO]-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
5WWP
 
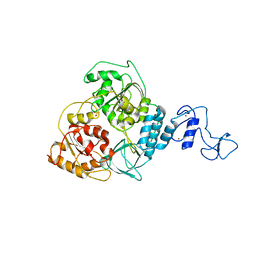 | | Crystal structure of Middle East respiratory syndrome coronavirus helicase (MERS-CoV nsp13) | | Descriptor: | ORF1ab, SULFATE ION, ZINC ION | | Authors: | Hao, W, Wojdyla, J.A, Zhao, R, Han, R, Das, R, Zlatev, I, Manoharan, M, Wang, M, Cui, S. | | Deposit date: | 2017-01-03 | | Release date: | 2017-07-05 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Middle East respiratory syndrome coronavirus helicase
PLoS Pathog., 13, 2017
|
|
2D41
 
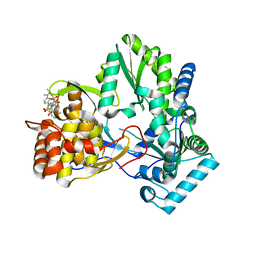 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor | | Descriptor: | 5'-ACETYL-4-{[(2,4-DIMETHYLPHENYL)SULFONYL]AMINO}-2,2'-BITHIOPHENE-5-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-05 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2D3Z
 
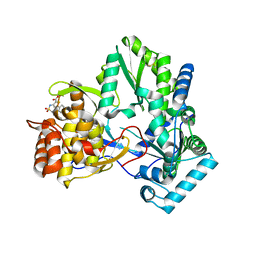 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-FLUOROPHENYL)-3-{[(4-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-04 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2D3U
 
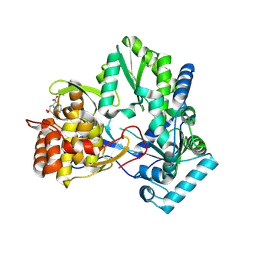 | | X-ray crystal structure of hepatitis C virus RNA dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-CYANOPHENYL)-3-{[(2-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-02 | | Release date: | 2006-08-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2FHG
 
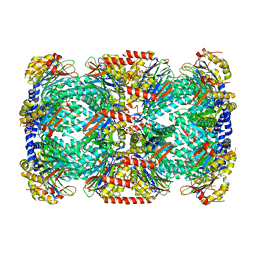 | | Crystal Structure of Mycobacterial Tuberculosis Proteasome | | Descriptor: | 20S proteasome, alpha and beta subunits, proteasome, ... | | Authors: | Hu, G, Lin, G, Wang, M, Dick, L, Xu, R.M, Nathan, C, Li, H. | | Deposit date: | 2005-12-23 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate.
Mol.Microbiol., 59, 2006
|
|
3U5U
 
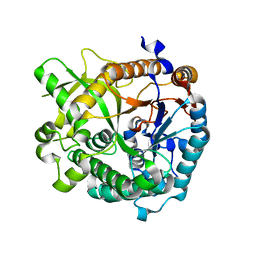 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
3U57
 
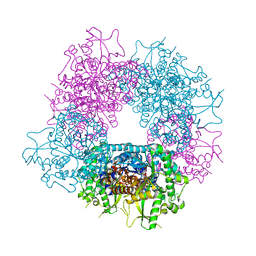 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | (2beta,7beta,16S,17R,19E,21beta)-21-(beta-D-glucopyranosyloxy)-2,7-dihydro-7,17-cyclosarpagan-17-yl acetate, CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
3U5Y
 
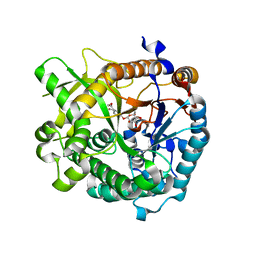 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | Descriptor: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase, Secologanin | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
7E9W
 
 | | The Crystal Structure of D-psicose-3-epimerase from Biortus. | | Descriptor: | D-psicose 3-epimerase, GLYCEROL, MANGANESE (II) ION | | Authors: | Wang, F, Xu, C, Qi, J, Zhang, M, Tian, F, Wang, M. | | Deposit date: | 2021-03-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of D-psicose-3-epimerase from Biortus.
To Be Published
|
|
3JUI
 
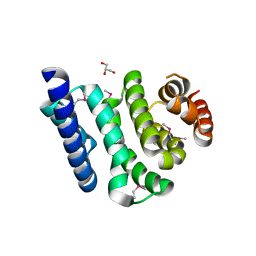 | | Crystal Structure of the C-terminal Domain of Human Translation Initiation Factor eIF2B epsilon Subunit | | Descriptor: | GLYCEROL, Translation initiation factor eIF-2B subunit epsilon | | Authors: | Wei, J, Xu, H, Zhang, C, Wang, M, Gao, F, Gong, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-10-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the C-terminal Domain of Human Translation Initiation Factor eIF2B epsilon Subunit
To be published
|
|
3ZJ7
 
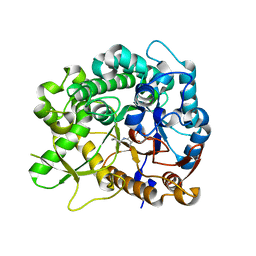 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-1 | | Descriptor: | (1R,2S,3S,4R,5R)-4-(cyclohexylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
3MDD
 
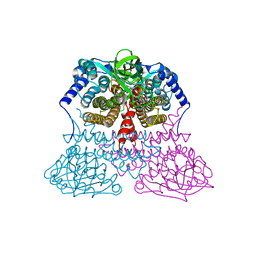 | |
3ZJ8
 
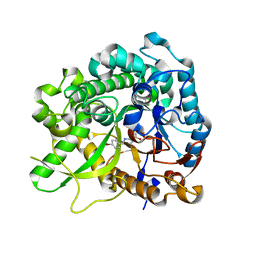 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-2 | | Descriptor: | (1R,2S,3S,4R,5R)-4-[(4-bromophenyl)methylamino]-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | Authors: | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
4A3Y
 
 | | Crystal structure of Raucaffricine glucosidase from ajmaline biosynthesis pathway | | Descriptor: | GLYCEROL, RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, SULFATE ION | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Barleben, L, Rajendran, C, Lin, H, Stoeckigt, J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity.
Acs Chem.Biol., 7, 2012
|
|
3GYR
 
 | | Structure of Phenoxazinone synthase from Streptomyces antibioticus reveals a new type 2 copper center. | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, GLYCEROL, ... | | Authors: | Smith, A.W, Camara-Artigas, A, Wang, M, Francisco, W.A, Allen, J.P. | | Deposit date: | 2009-04-05 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Phenoxazinone Synthase from Streptomyces Antibioticus Reveals a New Type 2 Copper Center.
Biochemistry, 45, 2006
|
|
3MDE
 
 | |
7CBO
 
 | | Crystal structure of beta-N-acetylhexosaminidase Am0868 from Akkermansia muciniphila in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL, ... | | Authors: | Xu, W, Wang, M, Zhang, M. | | Deposit date: | 2020-06-13 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and biochemical analyses of beta-N-acetylhexosaminidase Am0868 from Akkermansia muciniphila involved in mucin degradation.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
7CBN
 
 | |
7CGC
 
 | |
7CGD
 
 | | Silver-bound E.coli malate dehydrogenase | | Descriptor: | Malate dehydrogenase, SILVER ION | | Authors: | Wang, H, Wang, M, Sun, H. | | Deposit date: | 2020-07-01 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Atomic differentiation of silver binding preference in protein targets: Escherichia coli malate dehydrogenase as a paradigm.
Chem Sci, 11, 2020
|
|
6LHE
 
 | | Crystal Structure of Gold-bound NDM-1 | | Descriptor: | GOLD ION, Metallo-beta-lactamase type 2, SULFATE ION | | Authors: | Wang, H, Sun, H, Wang, M. | | Deposit date: | 2019-12-07 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.206 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
3VTP
 
 | | HIV fusion inhibitor MT-C34 | | Descriptor: | Transmembrane protein gp41 | | Authors: | Yao, X, Waltersperger, S, Wang, M, Cui, S. | | Deposit date: | 2012-06-02 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The M-T hook structure is critical for design of HIV-1 fusion inhibitors.
J.Biol.Chem., 287, 2012
|
|
4FIU
 
 | | The structure of hemagglutinin of H16 subtype influenza virus with V327G mutation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Lu, X, Shi, Y, Gao, F, Xiao, H, Wang, M, Qi, J, Gao, G.F. | | Deposit date: | 2012-06-11 | | Release date: | 2012-11-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Insights into Avian Influenza Virus Pathogenicity: the Hemagglutinin Precursor HA0 of Subtype H16 Has an Alpha-Helix Structure in Its Cleavage Site with Inefficient HA1/HA2 Cleavage.
J.Virol., 86, 2012
|
|
6LI6
 
 | | Crystal structure of MCR-1-S treated by Au(PEt3)Cl | | Descriptor: | GOLD ION, Probable phosphatidylethanolamine transferase Mcr-1, TRIETHYLPHOSPHANE | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
