1ZV8
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | ACETATE ION, CACODYLATE ION, E2 glycoprotein, ... | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
1ZV7
 
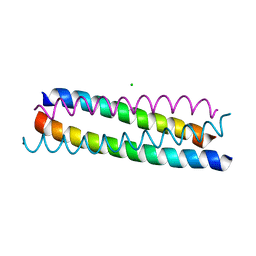 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | CHLORIDE ION, spike glycoprotein | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
4ZRD
 
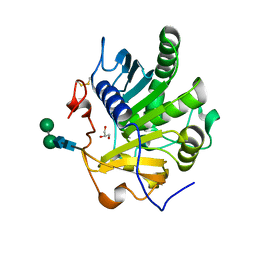 | | Crystal structure of SMG1 F278N mutant | | Descriptor: | GLYCEROL, LIP1, secretory lipase (Family 3), ... | | Authors: | Xu, J, Xu, H, Hou, S, Liu, J. | | Deposit date: | 2015-05-12 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of product-bound SMG1 lipase: active site gating implications.
Febs J., 282, 2015
|
|
4ZRE
 
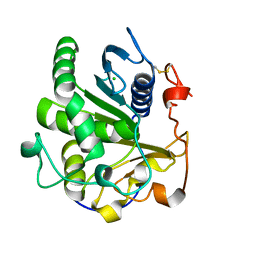 | | Crystal structure of SMG1 F278D mutant | | Descriptor: | CHLORIDE ION, LIP1, secretory lipase (Family 3), ... | | Authors: | Xu, J, Xu, H, Hou, S, Liu, J. | | Deposit date: | 2015-05-12 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of product-bound SMG1 lipase: active site gating implications.
Febs J., 282, 2015
|
|
4EJQ
 
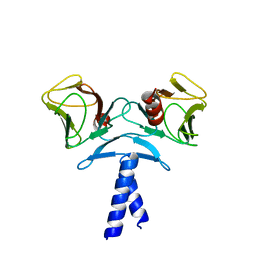 | | Crystal structure of KIF1A C-CC1-FHA | | Descriptor: | Kinesin-like protein KIF1A | | Authors: | Huo, L, Yue, Y, Ren, J, Yu, J, Liu, J, Yu, Y, Ye, F, Xu, T, Zhang, M, Feng, W. | | Deposit date: | 2012-04-06 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | The CC1-FHA Tandem as a Central Hub for Controlling the Dimerization and Activation of Kinesin-3 KIF1A
Structure, 20, 2012
|
|
3BDU
 
 | | Crystal structure of protein Q6D8G1 at the resolution 1.9 A. Northeast Structural Genomics Consortium target EwR22A. | | Descriptor: | Putative lipoprotein | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Fang, Y, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-15 | | Release date: | 2007-11-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of protein Q6D8G1 at the resolution 1.9 A.
To be Published
|
|
3BU2
 
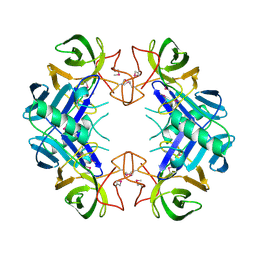 | | Crystal structure of a tRNA-binding protein from Staphylococcus saprophyticus subsp. saprophyticus. Northeast Structural Genomics Consortium target SyR77 | | Descriptor: | Putative tRNA-binding protein | | Authors: | Seetharaman, J, Su, M, Forouhar, F, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xia, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-12-31 | | Release date: | 2008-01-22 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a tRNA-binding protein from Staphylococcus saprophyticus subsp. saprophyticus.
To be Published
|
|
3GT0
 
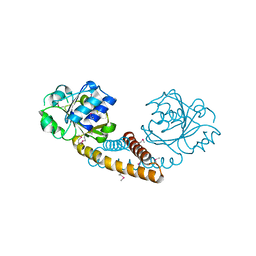 | | Crystal structure of pyrroline 5-carboxylate reductase from Bacillus cereus. Northeast Structural Genomics Consortium Target BcR38B | | Descriptor: | Pyrroline-5-carboxylate reductase | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Shastry, R, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-21 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Northeast Structural Genomics Consortium Target BcR38B
To be published
|
|
3C0B
 
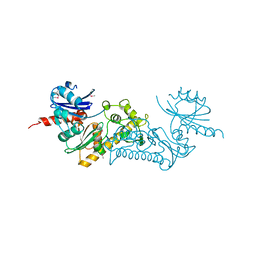 | | Crystal structure of the conserved archaeal protein Q6M145. Northeast Structural Genomics Consortium target MrR63 | | Descriptor: | CALCIUM ION, Conserved archaeal protein Q6M145 | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-19 | | Release date: | 2008-02-26 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of the conserved archaeal protein Q6M145.
To be Published
|
|
3C37
 
 | | X-ray structure of the putative Zn-dependent peptidase Q74D82 at the resolution 1.7 A. Northeast Structural Genomics Consortium target GsR143A | | Descriptor: | DI(HYDROXYETHYL)ETHER, Peptidase, M48 family, ... | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Vorobiev, S.M, Forouhar, F, Wang, D, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-27 | | Release date: | 2008-02-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the putative Zn-dependent peptidase Q74D82 at the resolution 1.7 A.
To be Published
|
|
2O18
 
 | | Crystal structure of a Thiamine biosynthesis lipoprotein apbE, NorthEast Strcutural Genomics target ER559 | | Descriptor: | CALCIUM ION, Thiamine biosynthesis lipoprotein apbE | | Authors: | Seetharaman, J, Su, M, Wang, D, Fang, Y, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a Thiamine biosynthesis lipoprotein apbE
To be Published
|
|
3C0D
 
 | | Crystal structure of the putative nitrite reductase NADPH (small subunit) oxidoreductase protein Q87HB1. Northeast Structural Genomics Consortium target VpR162 | | Descriptor: | Putative nitrite reductase NADPH (Small subunit) oxidoreductase protein | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Vorobiev, S.M, Wang, D, Fang, Y, Owens, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-19 | | Release date: | 2008-03-04 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the putative nitrite reductase NADPH (small subunit) oxidoreductase protein Q87HB1.
To be Published
|
|
2O8S
 
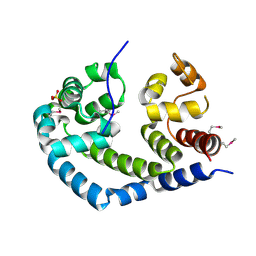 | | X-ray Crystal Structure of Protein AGR_C_984 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR120. | | Descriptor: | AGR_C_984p, SULFATE ION | | Authors: | Seetharaman, J, Forouhar, F, Su, M, Zhao, L, Cunningham, K, Ma, L.-C, Janjua, H, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-12 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Hypothetical Protein AGR_C_984 from Agrobacterium tumefaciens, Northeast Structural Genomics Target AtR120.
To be Published
|
|
2OBK
 
 | | X-Ray structure of the putative Se binding protein from Pseudomonas fluorescens. Northeast Structural Genomics Consortium target PlR6. | | Descriptor: | SelT/selW/selH selenoprotein domain | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-19 | | Release date: | 2007-01-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-Ray structure of the putative Se binding protein from Pseudomonas fluorescens
To be Published
|
|
2NVV
 
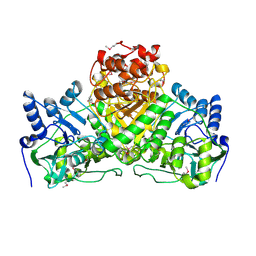 | | Crystal Structure of the Putative Acetyl-CoA hydrolase/transferase PG1013 from Porphyromonas gingivalis, Northeast Structural Genomics Target PgR16. | | Descriptor: | Acetyl-CoA hydrolase/transferase family protein, ZINC ION | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Yong, W, Ho, C.K, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-13 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Putative Acetyl-CoA hydrolase/transferase PG1013 from Porphyromonas gingivalis, Northeast Structural Genomics Target PgR16
To be Published
|
|
2O0Q
 
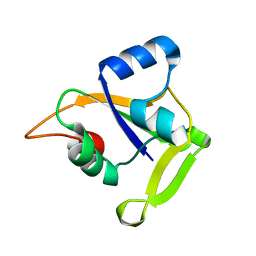 | | X-ray Crystal Structure of Protein CC0527 from Caulobacter crescentus. Northeast Structural Genomics Consortium Target CcR55 | | Descriptor: | Hypothetical protein CC0527 | | Authors: | Seetharaman, J, Su, M, Wang, D, Fang, Y, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Hypothetical Protein from Caulobacter Crescentus.
TO BE PUBLISHED
|
|
2O1E
 
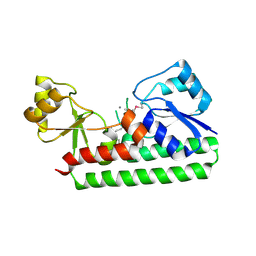 | | Crystal Structure of the Metal-dependent Lipoprotein YcdH from Bacillus subtilis, Northeast Structural Genomics Target SR583 | | Descriptor: | MANGANESE (II) ION, YcdH | | Authors: | Forouhar, F, Zhou, W, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.-C, Janjua, H, Xiao, R, Baran, M.C, Liu, J, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Metal-dependent Lipoprotein YcdH from Bacillus subtilis, Northeast Structural Genomics Target SR583
To be Published
|
|
3CFU
 
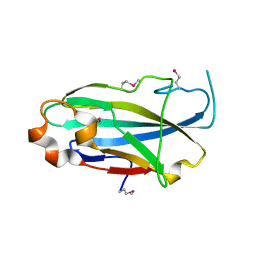 | | Crystal structure of the yjhA protein from Bacillus subtilis. Northeast Structural Genomics Consortium target SR562 | | Descriptor: | Uncharacterized lipoprotein yjhA | | Authors: | Vorobiev, S.M, Chen, Y, Kuzin, A.P, Seetharaman, J, Forouhar, F, Zhao, L, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Swapna, G, Huang, J.Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the yjhA protein from Bacillus subtilis.
To be Published
|
|
3CET
 
 | | Crystal structure of the pantheonate kinase-like protein Q6M145 at the resolution 1.8 A. Northeast Structural Genomics Consortium target MrR63 | | Descriptor: | Conserved archaeal protein, SULFATE ION | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Forouhar, F, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, J.F, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the pantheonate kinase-like protein Q6M145 at the resolution 1.8 A. Northeast Structural Genomics Consortium target MrR63.
To be Published
|
|
2O1M
 
 | | Crystal structure of the probable amino-acid ABC transporter extracellular-binding protein ytmK from Bacillus subtilis. Northeast Structural Genomics Consortium target SR572 | | Descriptor: | Probable amino-acid ABC transporter extracellular-binding protein ytmK, SULFATE ION | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-29 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the probable amino-acid ABC transporter extracellular-binding protein ytmK from Bacillus subtilis. Northeast Structural Genomics Consortium target SR572.
To be Published
|
|
2O14
 
 | | X-Ray Crystal Structure of Protein YXIM_BACsu from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR595 | | Descriptor: | Hypothetical protein yxiM, MANGANESE (II) ION, SULFATE ION | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray structure of the hypothetical protein YXIM_BACsu from Bacillus subtilis.
To be Published
|
|
2O0P
 
 | | X-ray Crystal Structure of Protein CC0527 (V27M / L66M double mutant) from Caulobacter crescentus. Northeast Structural Genomics Consortium Target CcR55. | | Descriptor: | Hypothetical protein CC0527 | | Authors: | Seetharaman, J, Su, M, Wang, D, Fang, Y, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-27 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Hypothetical Protein from Caulobacter Crescentus.
TO BE PUBLISHED
|
|
3D0W
 
 | | Crystal structure of YflH protein from Bacillus subtilis. Northeast Structural Genomics Consortium target SR326 | | Descriptor: | YflH protein | | Authors: | Seetharaman, J, Kuzin, A.P, Neely, H, Forouhar, F, Min, S, Zhao, L, Fang, Y, Owens, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-02 | | Release date: | 2008-05-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of YflH protein from Bacillus subtilis.
To be Published
|
|
3CER
 
 | | Crystal structure of the exopolyphosphatase-like protein Q8G5J2. Northeast Structural Genomics Consortium target BlR13 | | Descriptor: | Possible exopolyphosphatase-like protein, SULFATE ION | | Authors: | Kuzin, A.P, Su, M, Chen, Y, Neely, H, Seetharaman, J, Shastry, R, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-04-01 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the exopolyphosphatase-like protein Q8G5J2. Northeast Structural Genomics Consortium target BlR13.
To be Published
|
|
2P3Y
 
 | | Crystal structure of VPA0735 from Vibrio parahaemolyticus. NorthEast Structural Genomics target VpR109 | | Descriptor: | Hypothetical protein VPA0735 | | Authors: | Seetharaman, J, Neely, H, Forouhar, F, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xia, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-10 | | Release date: | 2007-03-27 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of VPA0735 from Vibrio parahaemolyticus.
To be Published
|
|
