6KV9
 
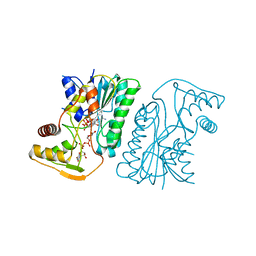 | | MoeE5 in complex with UDP-glucuronic acid and NAD | | Descriptor: | MoeE5, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Ko, T.-P, Liu, W, Sun, H, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-09-03 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of an antibiotic-synthesizing UDP-glucuronate 4-epimerase MoeE5 in complex with substrate.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
6KVC
 
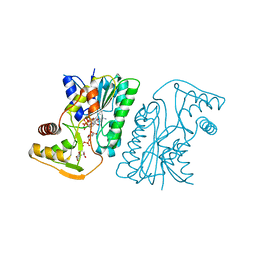 | | MoeE5 in complex with UDP-glucose and NAD | | Descriptor: | MoeE5, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Ko, T.-P, Liu, W, Sun, H, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of an antibiotic-synthesizing UDP-glucuronate 4-epimerase MoeE5 in complex with substrate.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
5XFY
 
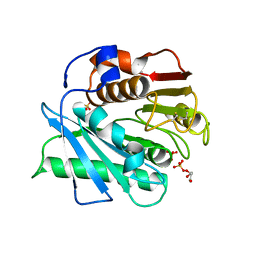 | | Crystal structure of a novel PET hydrolase S131A mutant from Ideonella sakaiensis 201-F6 | | Descriptor: | GLYCEROL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
5XG0
 
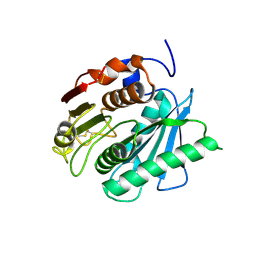 | | Crystal structure of a novel PET hydrolase from Ideonella sakaiensis 201-F6 | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
5XFZ
 
 | | Crystal structure of a novel PET hydrolase R103G/S131A mutant from Ideonella sakaiensis 201-F6 | | Descriptor: | GLYCEROL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
5XH2
 
 | | Crystal structure of a novel PET hydrolase R103G/S131A mutant in complex with pNP from Ideonella sakaiensis 201-F6 | | Descriptor: | P-NITROPHENOL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
5XO8
 
 | | Crystal structure of a novel ZEN lactonase mutant with ligand Z | | Descriptor: | (3S,11E)-14,16-dihydroxy-3-methyl-3,4,5,6,9,10-hexahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Lactonase for protein | | Authors: | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-05-27 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
5XH3
 
 | | Crystal structure of a novel PET hydrolase R103G/S131A mutant in complex with HEMT from Ideonella sakaiensis 201-F6 | | Descriptor: | GLYCEROL, O 4-(2-hydroxyethyl) O 1-methyl benzene-1,4-dicarboxylate, Poly(ethylene terephthalate) hydrolase, ... | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
6LDL
 
 | | Crystal structure of CYP116B46-N(20-445) from Tepidiphilus thermophilus in complex with HEME | | Descriptor: | BICINE, Cytochrome P450, GLYCEROL, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-11-21 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase.
Nat Commun, 11, 2020
|
|
3KP2
 
 | |
3KP4
 
 | |
3KP5
 
 | |
3KP6
 
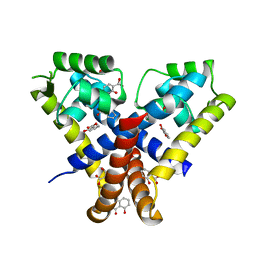 | | Staphylococcus epidermidis TcaR in complex with salicylate | | Descriptor: | 2-HYDROXYBENZOIC ACID, CACODYLATE ION, Transcriptional regulator TcaR | | Authors: | Chang, Y.M, Chen, C.K, Yeh, Y.J, Wang, A.H. | | Deposit date: | 2009-11-15 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural study of TcaR and its complexes with multiple antibiotics from Staphylococcus epidermidis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KP3
 
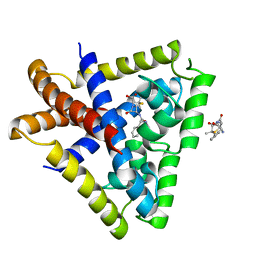 | | Staphylococcus epidermidis in complex with ampicillin | | Descriptor: | (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, Transcriptional regulator TcaR | | Authors: | Chang, Y.M, Chen, C.K, Wang, A.H. | | Deposit date: | 2009-11-15 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural study of TcaR and its complexes with multiple antibiotics from Staphylococcus epidermidis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3LGZ
 
 | | Crystal structure of dehydrosqualene synthase Y129A from S. aureus complexed with presqualene pyrophosphate | | Descriptor: | Dehydrosqualene synthase, MAGNESIUM ION, {(1R,2R,3R)-2-[(3E)-4,8-dimethylnona-3,7-dien-1-yl]-2-methyl-3-[(1E,5E)-2,6,10-trimethylundeca-1,5,9-trien-1-yl]cyclopropyl}methyl trihydrogen diphosphate | | Authors: | Lin, F.-Y, Liu, Y.-L, Liu, C.-I, Wang, A.H.J, Oldfield, E. | | Deposit date: | 2010-01-21 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Mechanism of action and inhibition of dehydrosqualene synthase.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4W4S
 
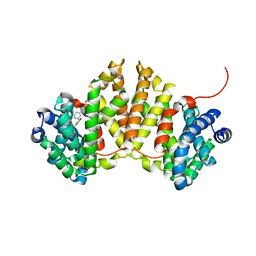 | | Crystal structure of ent-kaurene synthase BJKS from bradyrhizobium japonicum in complex with BPH-629 | | Descriptor: | Uncharacterized protein blr2150, [2-(3-DIBENZOFURAN-4-YL-PHENYL)-1-HYDROXY-1-PHOSPHONO-ETHYL]-PHOSPHONIC ACID | | Authors: | Liu, W, Zheng, Y, Huang, C.H, Guo, R.T. | | Deposit date: | 2014-08-15 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
5JSD
 
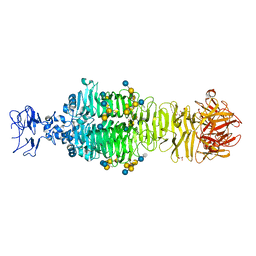 | | Crystal structure of phiAB6 tailspike in complex with five-repeated oligosaccharides of Acinetobacter baumannii surface polysaccharide | | Descriptor: | ACETIC ACID, MALONIC ACID, beta-D-galactopyranose-(1-3)-2-amino-2-deoxy-beta-D-galactopyranose-(1-3)-[5,7-bisacetamido-3,5,7,9-tetradeoxy-L-glycero-alpha-L-manno-non-2-ulopyranosonic acid-(2-6)-beta-D-glucopyranose-(1-6)]beta-D-galactopyranose-(1-3)-2-amino-2-deoxy-beta-D-galactopyranose-(1-3)-[beta-D-glucopyranose-(1-6)]beta-D-galactopyranose, ... | | Authors: | Lee, I.M, Tu, I.F, Huang, K.F, Wu, S.H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural basis for fragmenting the exopolysaccharide of Acinetobacter baumannii by bacteriophage Phi AB6 tailspike protein
Sci Rep, 7, 2017
|
|
5JS4
 
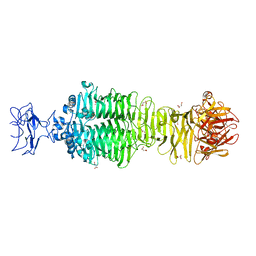 | | Crystal structure of phiAB6 tailspike | | Descriptor: | MALONIC ACID, phiAB6 tailspike | | Authors: | Lee, I.M, Tu, I.F, Huang, K.F, Wu, S.H. | | Deposit date: | 2016-05-07 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural basis for fragmenting the exopolysaccharide of Acinetobacter baumannii by bacteriophage Phi AB6 tailspike protein
Sci Rep, 7, 2017
|
|
5JSE
 
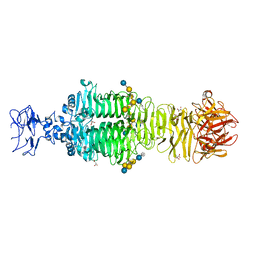 | | Crystal structure of phiAB6 tailspike in complex with three-repeated oligosaccharides of Acinetobacter baumannii surface polysaccharide | | Descriptor: | ACETIC ACID, MALONIC ACID, beta-D-galactopyranose-(1-3)-2-amino-2-deoxy-beta-D-galactopyranose-(1-3)-[5,7-bisacetamido-3,5,7,9-tetradeoxy-L-glycero-alpha-L-manno-non-2-ulopyranosonic acid-(2-6)-beta-D-glucopyranose-(1-6)]beta-D-galactopyranose-(1-3)-2-amino-2-deoxy-beta-D-galactopyranose-(1-3)-[beta-D-glucopyranose-(1-6)]beta-D-galactopyranose, ... | | Authors: | Lee, I.M, Tu, I.F, Huang, K.F, Wu, S.H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for fragmenting the exopolysaccharide of Acinetobacter baumannii by bacteriophage Phi AB6 tailspike protein
Sci Rep, 7, 2017
|
|
5KSJ
 
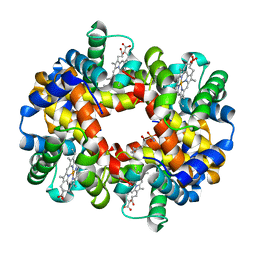 | | Crystal structure of deoxygenated hemoglobin in complex with Sphingosine phosphate | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Ahmed, M.H, Safo, M.K, Xia, Y. | | Deposit date: | 2016-07-08 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Insight of Sphingosine 1-Phosphate-Mediated Pathogenic Metabolic Reprogramming in Sickle Cell Disease.
Sci Rep, 7, 2017
|
|
5KSI
 
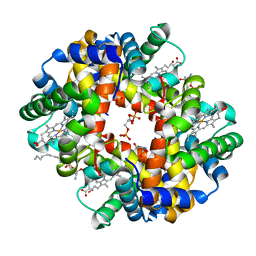 | | Crystal structure of deoxygenated hemoglobin in complex with sphingosine phosphate and 2,3-Bisphosphoglycerate | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Hemoglobin subunit alpha, ... | | Authors: | Ahmed, M.H, Safo, M.K, Xia, Y. | | Deposit date: | 2016-07-08 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Insight of Sphingosine 1-Phosphate-Mediated Pathogenic Metabolic Reprogramming in Sickle Cell Disease.
Sci Rep, 7, 2017
|
|
3PLN
 
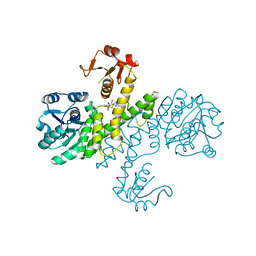 | | Crystal structure of Klebsiella pneumoniae UDP-glucose 6-dehydrogenase complexed with UDP-glucose | | Descriptor: | UDP-glucose 6-dehydrogenase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
3PJG
 
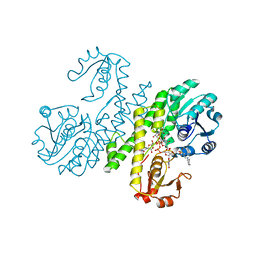 | | Crystal structure of UDP-glucose dehydrogenase from Klebsiella pneumoniae complexed with product UDP-glucuronic acid | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-10 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
3PID
 
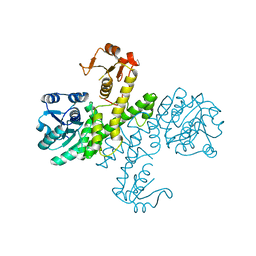 | | The apo-form UDP-glucose 6-dehydrogenase with a C-terminal six-histidine tag | | Descriptor: | UDP-glucose 6-dehydrogenase | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-06 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
3PLR
 
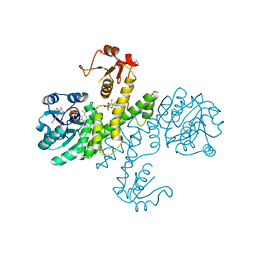 | | Crystal structure of Klebsiella pneumoniae UDP-glucose 6-dehydrogenase complexed with NADH and UDP-glucose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, UDP-glucose 6-dehydrogenase, URIDINE-5'-MONOPHOSPHATE | | Authors: | Chen, Y.-Y, Ko, T.-P, Lin, C.-H, Chen, W.-H, Wang, A.H.-J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance.
J.Struct.Biol., 175, 2011
|
|
