6UEP
 
 | |
6UEO
 
 | | Structure of A. thaliana TBP-AC mismatch DNA site | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*CP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TATA-box-binding protein 1 | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA mismatches reveal conformational penalties in protein-DNA recognition.
Nature, 587, 2020
|
|
6UER
 
 | |
6UEQ
 
 | | Structure of TBP bound to C-C mismatch containing TATA site | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*CP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*C)-3'), SULFATE ION, ... | | Authors: | Schumacher, M.A, Al-Hashimi, H. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DNA mismatches reveal conformational penalties in protein-DNA recognition.
Nature, 587, 2020
|
|
6NJQ
 
 | | Structure of TBP-Hoogsteen containing DNA complex | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*CP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*GP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TATA-box-binding protein 1 | | Authors: | Schumacher, M.A, Stelling, A. | | Deposit date: | 2019-01-04 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Infrared Spectroscopic Observation of a G-C+Hoogsteen Base Pair in the DNA:TATA-Box Binding Protein Complex Under Solution Conditions.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
8CVT
 
 | | Human 19S-20S proteasome, state SD2 | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 8, 26S proteasome complex subunit SEM1, ... | | Authors: | Zhao, J. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5A5B
 
 | | Structure of the 26S proteasome-Ubp6 complex | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Aufderheide, A, Beck, F, Stengel, F, Hartwig, M, Schweitzer, A, Pfeifer, G, Goldberg, A.L, Sakata, E, Baumeister, W, Foerster, F. | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-22 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structural Characterization of the Interaction of Ubp6 with the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1VTO
 
 | |
7QO5
 
 | | 26S proteasome Rpt1-RK -Ubp6-UbVS complex in the si state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-03-16 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
6WJN
 
 | |
6WJD
 
 | |
8USB
 
 | |
8USC
 
 | | Nub1/Fat10-processing human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 8, 26S proteasome complex subunit SEM1, ... | | Authors: | Arkinson, C, Gee, C.L, Martin, A. | | Deposit date: | 2023-10-27 | | Release date: | 2024-11-06 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | NUB1 traps unfolded FAT10 for ubiquitin-independent degradation by the 26S proteasome.
Nat.Struct.Mol.Biol., 2025
|
|
5GJQ
 
 | | Structure of the human 26S proteasome bound to USP14-UbAl | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Huang, X.L, Luan, B, Wu, J.P, Shi, Y.G. | | Deposit date: | 2016-07-01 | | Release date: | 2016-08-17 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | An atomic structure of the human 26S proteasome.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5GJR
 
 | | An atomic structure of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Huang, X.L, Luan, B, Wu, J.P, Shi, Y.G. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An atomic structure of the human 26S proteasome.
Nat. Struct. Mol. Biol., 23, 2016
|
|
3JCO
 
 | | Structure of yeast 26S proteasome in M1 state derived from Titan dataset | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3JCP
 
 | | Structure of yeast 26S proteasome in M2 state derived from Titan dataset | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3WHK
 
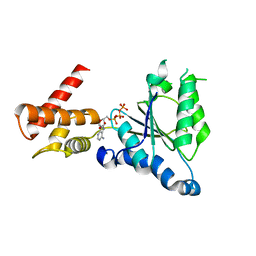 | | Crystal structure of PAN-Rpt5C chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Proteasome-activating nucleotidase, 26S protease regulatory subunit 6A | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WHL
 
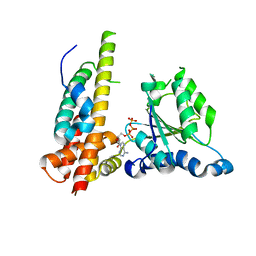 | | Crystal structure of Nas2 N-terminal domain complexed with PAN-Rpt5C chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Probable 26S proteasome regulatory subunit p27, Proteasome-activating nucleotidase, ... | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
5L4G
 
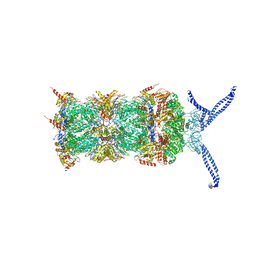 | | The human 26S proteasome at 3.9 A | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Schweitzer, A, Aufderheide, A, Rudack, T, Beck, F. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the human 26S proteasome at a resolution of 3.9 angstrom.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5LN3
 
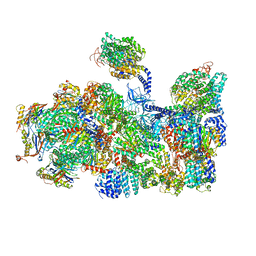 | | The human 26S Proteasome at 6.8 Ang. | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Schweitzer, A, Beck, F, Sakata, E, Unverdorben, P. | | Deposit date: | 2016-08-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular Details Underlying Dynamic Structures and Regulation of the Human 26S Proteasome.
Mol. Cell Proteomics, 16, 2017
|
|
5M32
 
 | | Human 26S proteasome in complex with Oprozomib | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Haselbach, D, Schrader, J, Lambrecht, F, Henneberg, F, Chari, A, Stark, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-07-05 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Long-range allosteric regulation of the human 26S proteasome by 20S proteasome-targeting cancer drugs.
Nat Commun, 8, 2017
|
|
4CR2
 
 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CR4
 
 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CR3
 
 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
