2KTR
 
 | | NMR structure of p62 PB1 dimer determined based on PCS | | Descriptor: | Sequestosome-1, TERBIUM(III) ION | | Authors: | Saio, T, Yokochi, M, Kumeta, H, Inagaki, F. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-07 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | PCS-based structure determination of protein-protein complexes
J.Biomol.Nmr, 46, 2010
|
|
2FFZ
 
 | |
2FGN
 
 | |
2HUC
 
 | |
7U9D
 
 | | Crystal Structure of Human Phosphatidylcholine Transfer Protein in Complex with PC(16:0/20:4) | | Descriptor: | GLYCEROL, Phosphatidylcholine transfer protein, [(2~{R})-1-hexadecanoyloxy-3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxy-propan-2-yl] (5~{Z},8~{Z},11~{Z},14~{Z})-icosa-5,8,11,14-tetraenoate | | Authors: | Druzak, S.A. | | Deposit date: | 2022-03-10 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Ligand dependent interaction between PC-TP and PPAR delta mitigates diet-induced hepatic steatosis in male mice.
Nat Commun, 14, 2023
|
|
2BV8
 
 | | The crystal structure of Phycocyanin from Gracilaria chilensis. | | Descriptor: | C-PHYCOCYANIN ALPHA SUBUNIT, C-PHYCOCYANIN BETA SUBUNIT, PHYCOCYANOBILIN, ... | | Authors: | Contreras-Martel, C, Martinez-Oyanedel, J, Poo-Caama, G, Bruna, C, Bunster, M. | | Deposit date: | 2005-06-23 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Structure at 2 A Resolution of Phycocyanin from Gracilaria Chilensis and the Energy Transfer Network in a Pc-Pc Complex.
Biophys.Chem., 125, 2007
|
|
1Q1O
 
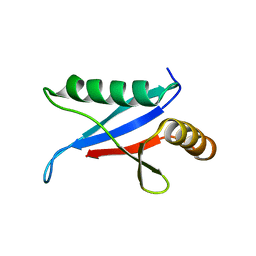 | | Solution Structure of the PB1 Domain of Cdc24p (Long Form) | | Descriptor: | Cell division control protein 24 | | Authors: | Yoshinaga, S, Kohjima, M, Ogura, K, Yokochi, M, Takeya, R, Ito, T, Sumimoto, H, Inagaki, F. | | Deposit date: | 2003-07-22 | | Release date: | 2003-10-14 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The PB1 domain and the PC motif-containing region are structurally similar protein binding modules
EMBO J., 22, 2003
|
|
1IPG
 
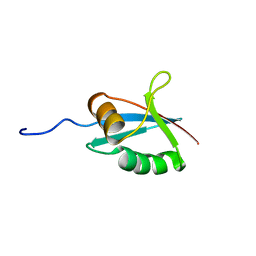 | | SOLUTION STRUCTURE OF THE PB1 DOMAIN OF BEM1P | | Descriptor: | BEM1 PROTEIN | | Authors: | Terasawa, H, Noda, Y, Ito, T, Hatanaka, H, Ichikawa, S, Ogura, K, Sumimoto, H, Inagaki, F. | | Deposit date: | 2001-05-14 | | Release date: | 2001-08-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and ligand recognition of the PB1 domain: a novel protein module binding to the PC motif.
EMBO J., 20, 2001
|
|
1IP9
 
 | | SOLUTION STRUCTURE OF THE PB1 DOMAIN OF BEM1P | | Descriptor: | BEM1 PROTEIN | | Authors: | Terasawa, H, Noda, Y, Ito, T, Hatanaka, H, Ichikawa, S, Ogura, K, Sumimoto, H, Inagaki, F. | | Deposit date: | 2001-04-26 | | Release date: | 2001-08-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and ligand recognition of the PB1 domain: a novel protein module binding to the PC motif.
EMBO J., 20, 2001
|
|
4ADY
 
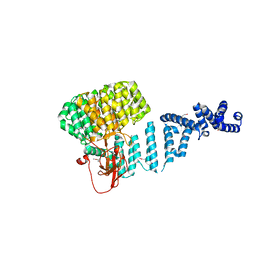 | | Crystal structure of 26S proteasome subunit Rpn2 | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN2 | | Authors: | Kulkarni, K, He, J, Da Fonseca, P.C.A, Krutauz, D, Glickman, M.H, Barford, D, Morris, E.P. | | Deposit date: | 2012-01-04 | | Release date: | 2012-03-14 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of the 26S Proteasome Subunit Rpn2 Reveals its Pc Repeat Domain as a Closed Toroid of Two Concentric Alpha-Helical Rings
Structure, 20, 2012
|
|
1NG8
 
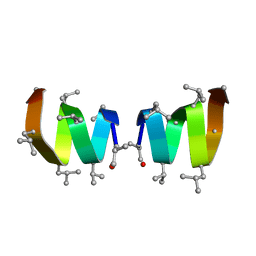 | |
1NT6
 
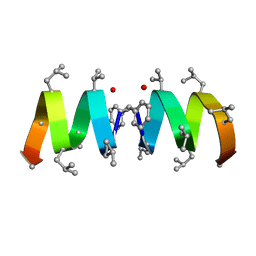 | | F1-Gramicidin C In Sodium Dodecyl Sulfate Micelles (NMR) | | Descriptor: | GRAMICIDIN C | | Authors: | Townsley, L.E, Fletcher, T.G, Hinton, J.F. | | Deposit date: | 2003-01-28 | | Release date: | 2003-02-11 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The Structure, Cation Binding, Transport, and Conductance of Gly15-Gramicidin a Incorporated Into Sds Micelles and Pc/Pg Vesicles.
Biochemistry, 42, 2003
|
|
1NT5
 
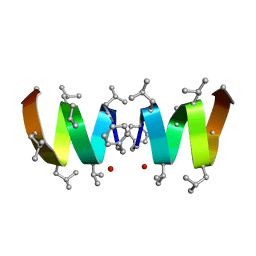 | | F1-Gramicidin A in Sodium Dodecyl Sulfate Micelles (NMR) | | Descriptor: | GRAMICIDIN A | | Authors: | Townsley, L.E, Fletcher, T.G, Hinton, J.F. | | Deposit date: | 2003-01-28 | | Release date: | 2003-02-11 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The Structure, Cation Binding, Transport, and Conductance of Gly15-Gramicidin a Incorporated Into Sds Micelles and Pc/Pg Vesicles.
Biochemistry, 42, 2003
|
|
5CUE
 
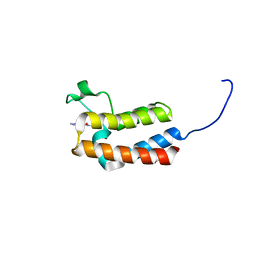 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with AGN-PC-04G0SS (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N,1-dimethyl-1H-pyrazole-4-carboxamide, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with AGN-PC-04G0SS (SGC - Diamond I04-1 fragment screening)
To be published
|
|
7SNW
 
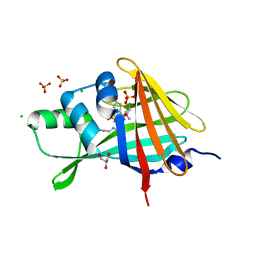 | | 1.80A Resolution Structure of NanoLuc Luciferase with Bound Inhibitor PC 16026576 | | Descriptor: | 2-(methoxycarbonyl)thiophene-3-sulfonic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.80A Resolution Structure of NanoLuc Luciferase with Bound Inhibitor PC 16026576
To be published
|
|
2MCP
 
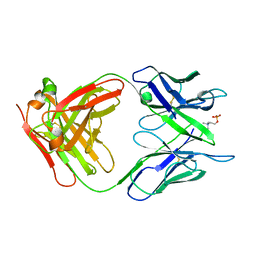 | |
1B09
 
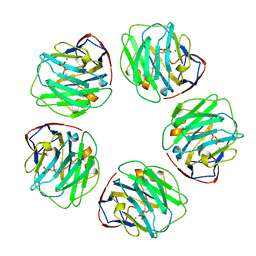 | |
1PDQ
 
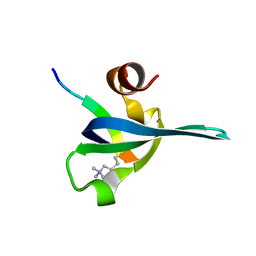 | | Polycomb chromodomain complexed with the histone H3 tail containing trimethyllysine 27. | | Descriptor: | Histone H3.3, Polycomb protein | | Authors: | Fischle, W, Wang, Y, Jacobs, S.A, Kim, Y, Allis, C.D, Khorasanizadeh, S. | | Deposit date: | 2003-05-20 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains
Genes Dev., 17, 2003
|
|
2RSE
 
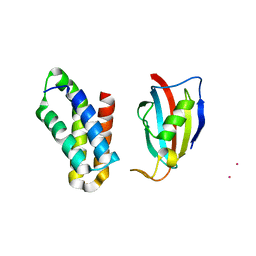 | | NMR structure of FKBP12-mTOR FRB domain-rapamycin complex structure determined based on PCS | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Serine/threonine-protein kinase mTOR, TERBIUM(III) ION | | Authors: | Kobashigawa, Y, Ushio, M, Saio, T, Inagaki, F. | | Deposit date: | 2012-01-25 | | Release date: | 2012-05-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Convenient method for resolving degeneracies due to symmetry of the magnetic susceptibility tensor and its application to pseudo contact shift-based protein-protein complex structure determination.
J.Biomol.Nmr, 53, 2012
|
|
6P3Q
 
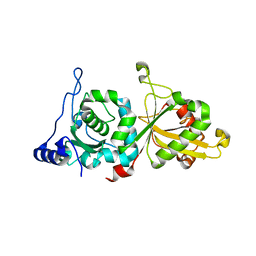 | | Calpain-5 (CAPN5) Protease Core (PC) | | Descriptor: | Calpain-5 | | Authors: | Velez, G, Sun, Y.J, Khan, S, Yang, J, Koster, H.J, Lokesh, G, Mahajan, V. | | Deposit date: | 2019-05-24 | | Release date: | 2020-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Unique Activation Mechanisms of a Non-classical Calpain and Its Disease-Causing Variants.
Cell Rep, 30, 2020
|
|
4ZCR
 
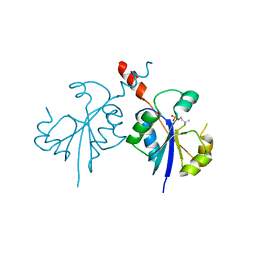 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase in complex with phosphocholine | | Descriptor: | Cholinephosphate cytidylyltransferase, PHOSPHOCHOLINE | | Authors: | Guca, E, Hoh, F, Guichou, J.-F, Cerdan, R. | | Deposit date: | 2015-04-16 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants of the catalytic mechanism of Plasmodium CCT, a key enzyme of malaria lipid biosynthesis.
Sci Rep, 8, 2018
|
|
4CLV
 
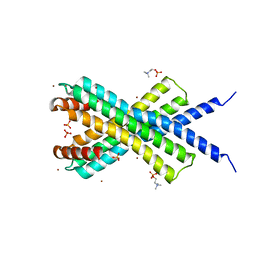 | | Crystal Structure of dodecylphosphocholine-solubilized NccX from Cupriavidus metallidurans 31A | | Descriptor: | NICKEL-COBALT-CADMIUM RESISTANCE PROTEIN NCCX, PHOSPHATE ION, PHOSPHOCHOLINE, ... | | Authors: | Legrand, P, Girard, E, Petit-Hartlein, I, Maillard, A.P, Coves, J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-10-01 | | Last modified: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The X-Ray Structure of Nccx from Cupriavidus Metallidurans 31A Illustrates Potential Dangers of Detergent Solubilization When Generating and Interpreting Crystal Structures of Membrane Proteins.
J.Biol.Chem., 289, 2014
|
|
1H8P
 
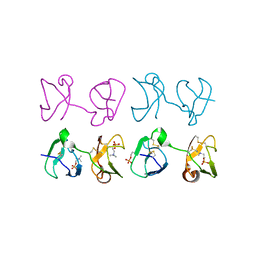 | | Bull seminal plasma PDC-109 fibronectin type II module | | Descriptor: | PHOSPHOCHOLINE, SEMINAL PLASMA PROTEIN PDC-109 | | Authors: | Wah, D.A, Fernandez-Tornero, C, Calvete, J.J, Romero, A. | | Deposit date: | 2001-02-14 | | Release date: | 2002-04-12 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Sperm Coating Mechanism from the 1.8 A Crystal Structure of Pdc-109-Phosphorylcholine Complex
Structure, 10, 2002
|
|
2GOH
 
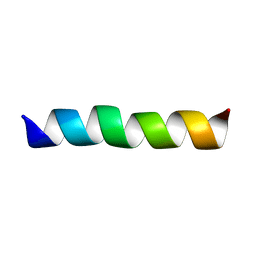 | | Three-dimensional Structure of the Trans-membrane Domain of Vpu from HIV-1 in Aligned Phospholipid Bicelles | | Descriptor: | VPU protein | | Authors: | Park, S.H, De Angelis, A.A, Nevzorov, A.A, Wu, C.H, Opella, S.J. | | Deposit date: | 2006-04-12 | | Release date: | 2006-08-08 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure of the Transmembrane Domain of Vpu from HIV-1 in Aligned Phospholipid Bicelles.
Biophys.J., 91, 2006
|
|
2GOF
 
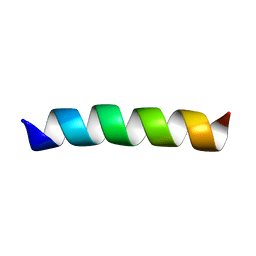 | | Three-dimensional structure of the trans-membrane domain of Vpu from HIV-1 in aligned phospholipid bicelles | | Descriptor: | VPU protein | | Authors: | Park, S.H, De Angelis, A.A, Nevzorov, A.A, Wu, C.H, Opella, S.J. | | Deposit date: | 2006-04-12 | | Release date: | 2006-08-08 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Structure of the Transmembrane Domain of Vpu from HIV-1 in Aligned Phospholipid Bicelles.
Biophys.J., 91, 2006
|
|
