1CEL
 
 | |
2CEL
 
 | | ACTIVE-SITE MUTANT E212Q DETERMINED AT PH 6.0 WITH NO LIGAND BOUND IN THE ACTIVE SITE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1996-08-24 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 264, 1996
|
|
3CEL
 
 | | ACTIVE-SITE MUTANT E212Q DETERMINED AT PH 6.0 WITH CELLOBIOSE BOUND IN THE ACTIVE SITE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1996-08-24 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 264, 1996
|
|
4CEL
 
 | | ACTIVE-SITE MUTANT D214N DETERMINED AT PH 6.0 WITH NO LIGAND BOUND IN THE ACTIVE SITE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1996-08-24 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 264, 1996
|
|
1EG1
 
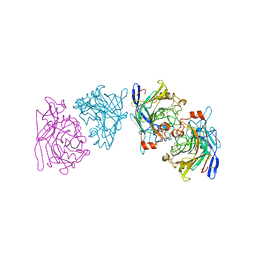 | | ENDOGLUCANASE I FROM TRICHODERMA REESEI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Kleywegt, G.J, Zou, J.-Y, Jones, T.A. | | Deposit date: | 1996-11-26 | | Release date: | 1997-08-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The crystal structure of the catalytic core domain of endoglucanase I from Trichoderma reesei at 3.6 A resolution, and a comparison with related enzymes.
J.Mol.Biol., 272, 1997
|
|
1OVW
 
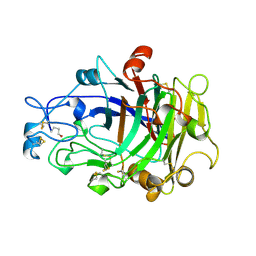 | | ENDOGLUCANASE I COMPLEXED WITH NON-HYDROLYSABLE SUBSTRATE ANALOGUE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-thio-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-1,4-dithio-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Sulzenbacher, G, Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-10-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Fusarium oxysporum endoglucanase I with a nonhydrolyzable substrate analogue: substrate distortion gives rise to the preferred axial orientation for the leaving group.
Biochemistry, 35, 1996
|
|
5CEL
 
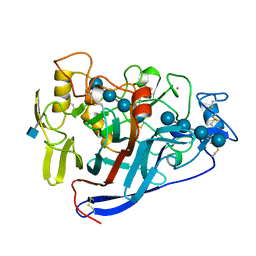 | | CBH1 (E212Q) CELLOTETRAOSE COMPLEX | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
7CEL
 
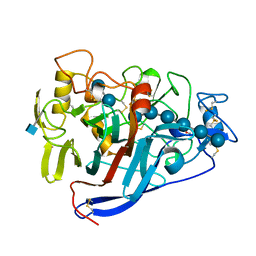 | | CBH1 (E217Q) IN COMPLEX WITH CELLOHEXAOSE AND CELLOBIOSE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
6CEL
 
 | | CBH1 (E212Q) CELLOPENTAOSE COMPLEX | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
3OVW
 
 | | ENDOGLUCANASE I NATIVE STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the endoglucanase I from Fusarium oxysporum: native, cellobiose, and 3,4-epoxybutyl beta-D-cellobioside-inhibited forms, at 2.3 A resolution.
Biochemistry, 36, 1997
|
|
2OVW
 
 | | ENDOGLUCANASE I COMPLEXED WITH CELLOBIOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Sulzenbacher, G, Davies, G.J, Schulein, M. | | Deposit date: | 1997-04-04 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the endoglucanase I from Fusarium oxysporum: native, cellobiose, and 3,4-epoxybutyl beta-D-cellobioside-inhibited forms, at 2.3 A resolution.
Biochemistry, 36, 1997
|
|
4OVW
 
 | | ENDOGLUCANASE I COMPLEXED WITH EPOXYBUTYL CELLOBIOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(beta-D-glucopyranosyloxy)-2,2-dihydroxybutyl propanoate, ENDOGLUCANASE I | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the endoglucanase I from Fusarium oxysporum: native, cellobiose, and 3,4-epoxybutyl beta-D-cellobioside-inhibited forms, at 2.3 A resolution.
Biochemistry, 36, 1997
|
|
2A39
 
 | | HUMICOLA INSOLENS ENDOCELLULASE EGI NATIVE STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Sulzenbacher, G, Mackenzie, L, Withers, S.G, Divne, C, Jones, T.A, Woldike, H.F, Schulein, M. | | Deposit date: | 1998-01-30 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 A resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate.
Biochem.J., 335, 1998
|
|
1A39
 
 | | HUMICOLA INSOLENS ENDOCELLULASE EGI S37W, P39W DOUBLE-MUTANT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Ducros, V, Lewis, R.J, Borchert, T.V, Schulein, M. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oligosaccharide specificity of a family 7 endoglucanase: insertion of potential sugar-binding subsites.
J.Biotechnol., 57, 1997
|
|
1DYM
 
 | | Humicola insolens Endocellulase Cel7B (EG 1) E197A Mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Moraz, O, Driguez, H, Schulein, M. | | Deposit date: | 2000-02-03 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 A resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate.
Biochem.J., 335 ( Pt 2), 1998
|
|
1DY4
 
 | | CBH1 IN COMPLEX WITH S-PROPRANOLOL | | Descriptor: | 1-(ISOPROPYLAMINO)-3-(1-NAPHTHYLOXY)-2-PROPANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Stahlberg, J, Henriksson, H, Divne, C, Isaksson, R, Pettersson, G, Johansson, G, Jones, T.A. | | Deposit date: | 2000-01-26 | | Release date: | 2000-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Enantiomer Binding and Separation of a Common Beta-Blocker: Crystal Structure of Cellobiohydrolase Cel7A with Bound (S)-Propranolol at 1.9 A Resolution
J.Mol.Biol., 305, 2001
|
|
1EGN
 
 | | CELLOBIOHYDROLASE CEL7A (E223S, A224H, L225V, T226A, D262G) MUTANT | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE CEL7A, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2000-02-16 | | Release date: | 2001-05-16 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering of a glycosidase Family 7 cellobiohydrolase to more alkaline pH optimum: the pH behaviour of Trichoderma reesei Cel7A and its E223S/ A224H/L225V/T226A/D262G mutant.
Biochem.J., 356, 2001
|
|
1GPI
 
 | |
1H46
 
 | | The catalytic module of Cel7D from Phanerochaete chrysosporium as a chiral selector: Structural studies of its complex with the b-blocker (R)-propranolol | | Descriptor: | (1E,2R)-1-(ISOPROPYLIMINO)-3-(1-NAPHTHYLOXY)PROPAN-2-OL, 2-acetamido-2-deoxy-beta-D-glucopyranose, EXOGLUCANASE I | | Authors: | Munoz, I.G, Mowbray, S.L, Stahlberg, J. | | Deposit date: | 2002-10-03 | | Release date: | 2003-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The Catalytic Module of Cel7D from Phanerochaete Chrysosporium as a Chiral Selector: Structural Studies of its Complex with the Beta Blocker (R)-Propranolol
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1Q2B
 
 | | CELLOBIOHYDROLASE CEL7A WITH DISULPHIDE BRIDGE ADDED ACROSS EXO-LOOP BY MUTATIONS D241C AND D249C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, EXOCELLOBIOHYDROLASE I | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2003-07-24 | | Release date: | 2003-11-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D.
J.Mol.Biol., 333, 2003
|
|
1Q2E
 
 | | CELLOBIOHYDROLASE CEL7A WITH LOOP DELETION 245-252 AND BOUND NON-HYDROLYSABLE CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EXOCELLOBIOHYDROLASE I, ... | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2003-07-24 | | Release date: | 2003-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D
J.Mol.Biol., 333, 2003
|
|
1OJJ
 
 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7BE197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
1OJK
 
 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7BE197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, GLYCEROL, ... | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
1OJI
 
 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7B E197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, GLYCEROL | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
1Q9H
 
 | | 3-Dimensional structure of native Cel7A from Talaromyces emersonii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, cellobiohydrolase I catalytic domain | | Authors: | Grassick, A, Thompson, R, Murray, P.G, Collins, C.M, Byrnes, L, Tuohy, M.G, Birrane, G, Higgins, T.M. | | Deposit date: | 2003-08-25 | | Release date: | 2004-11-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Three-dimensional structure of a thermostable native cellobiohydrolase, CBH IB, and molecular characterization of the cel7 gene from the filamentous fungus, Talaromyces emersonii
Eur.J.Biochem., 271, 2004
|
|
