5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | Authors: | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2015-11-25 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
7OVZ
 
 | |
1A93
 
 | | NMR SOLUTION STRUCTURE OF THE C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MAX PROTEIN, MYC PROTO-ONCOGENE PROTEIN | | Authors: | Lavigne, P, Crump, M.P, Gagne, S.M, Hodges, R.S, Kay, C.M, Sykes, B.D. | | Deposit date: | 1998-04-15 | | Release date: | 1998-10-21 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper.
J.Mol.Biol., 281, 1998
|
|
1R05
 
 | | Solution Structure of Max B-HLH-LZ | | Descriptor: | Max protein | | Authors: | Sauv, S, Tremblay, L, Lavigne, P. | | Deposit date: | 2003-09-19 | | Release date: | 2003-10-21 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of a mutant of the Max b/HLH/LZ free of DNA: insights into the specific and reversible DNA binding mechanism of dimeric transcription factors
J.Mol.Biol., 342, 2004
|
|
1HLO
 
 | | THE CRYSTAL STRUCTURE OF AN INTACT HUMAN MAX-DNA COMPLEX: NEW INSIGHTS INTO MECHANISMS OF TRANSCRIPTIONAL CONTROL | | Descriptor: | DNA (5'-D(*AP*CP*CP*AP*CP*GP*TP*GP*GP*TP*G)-3'), DNA (5'-D(*CP*AP*CP*CP*AP*CP*GP*TP*GP*GP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR MAX) | | Authors: | Brownlie, P, Ceska, T.A, Lamers, M, Romier, C, Theo, H, Suck, D. | | Deposit date: | 1997-09-10 | | Release date: | 1997-10-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of an intact human Max-DNA complex: new insights into mechanisms of transcriptional control.
Structure, 5, 1997
|
|
2A93
 
 | | NMR SOLUTION STRUCTURE OF THE C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER, 40 STRUCTURES | | Descriptor: | C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER | | Authors: | Lavigne, P, Crump, M.P, Gagne, S.M, Hodges, R.S, Kay, C.M, Sykes, B.D. | | Deposit date: | 1998-06-09 | | Release date: | 1999-01-27 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper.
J.Mol.Biol., 281, 1998
|
|
3U5V
 
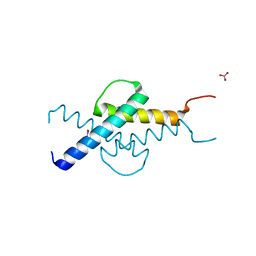 | | Crystal structure of Max-E47 | | Descriptor: | NITRATE ION, Protein max, Transcription factor E2-alpha chimera | | Authors: | Guarne, A, Ahmadpour, F, Gloyd, M. | | Deposit date: | 2011-10-11 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the minimalist max-e47 protein chimera.
Plos One, 7, 2012
|
|
6HZ2
 
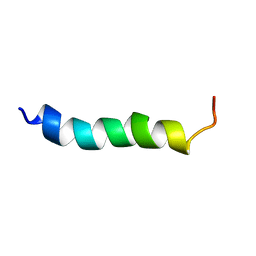 | |
1AN2
 
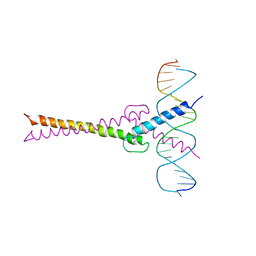 | | RECOGNITION BY MAX OF ITS COGNATE DNA THROUGH A DIMERIC B/HLH/Z DOMAIN | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*GP*GP*TP*CP*AP*CP*GP*TP*GP*AP*CP*C P*TP*AP*CP*AP*C)- 3'), PROTEIN (TRANSCRIPTION FACTOR MAX (TF MAX)) | | Authors: | Ferre-D'Amare, A.R, Prendergast, G.C, Ziff, E.B, Burley, S.K. | | Deposit date: | 1996-09-06 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain.
Nature, 363, 1993
|
|
6G6L
 
 | | The crystal structures of Human MYC:MAX bHLHZip complex | | Descriptor: | Myc proto-oncogene protein, Protein max, SULFATE ION | | Authors: | Allen, M.D, Zinzalla, G. | | Deposit date: | 2018-04-01 | | Release date: | 2019-04-10 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures and Nuclear Magnetic Resonance Studies of the Apo Form of the c-MYC:MAX bHLHZip Complex Reveal a Helical Basic Region in the Absence of DNA.
Biochemistry, 58, 2019
|
|
6G6J
 
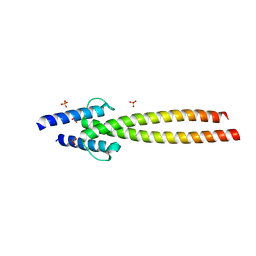 | | The crystal structures of Human MYC:MAX bHLHZip complex | | Descriptor: | Myc proto-oncogene protein, Protein max, SULFATE ION | | Authors: | Allen, M.D, Zinzalla, G. | | Deposit date: | 2018-04-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures and Nuclear Magnetic Resonance Studies of the Apo Form of the c-MYC:MAX bHLHZip Complex Reveal a Helical Basic Region in the Absence of DNA.
Biochemistry, 58, 2019
|
|
6G6K
 
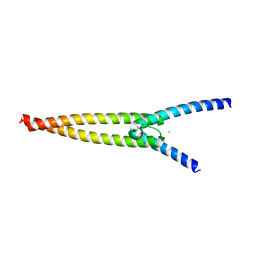 | | The crystal structures of Human MYC:MAX bHLHZip complex | | Descriptor: | CHLORIDE ION, Myc proto-oncogene protein, Protein max | | Authors: | Allen, M.D, Zinzalla, G. | | Deposit date: | 2018-04-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structures and Nuclear Magnetic Resonance Studies of the Apo Form of the c-MYC:MAX bHLHZip Complex Reveal a Helical Basic Region in the Absence of DNA.
Biochemistry, 58, 2019
|
|
1NKP
 
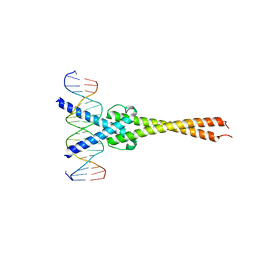 | | Crystal structure of Myc-Max recognizing DNA | | Descriptor: | 5'-D(*CP*GP*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*TP*C)-3', Max protein, Myc proto-oncogene protein | | Authors: | Nair, S.K, Burley, S.K. | | Deposit date: | 2003-01-03 | | Release date: | 2003-02-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structures of Myc-Max and Mad-Max recognizing DNA: Molecular bases of regulation by proto-oncogenic transcription factors
Cell(Cambridge,Mass.), 112, 2003
|
|
1NLW
 
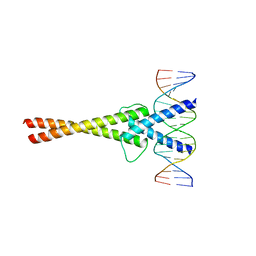 | | Crystal structure of Mad-Max recognizing DNA | | Descriptor: | 5'-D(*GP*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*TP*C)-3', MAD PROTEIN, MAX PROTEIN | | Authors: | Nair, S.K, Burley, S.K. | | Deposit date: | 2003-01-07 | | Release date: | 2003-02-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of Myc-Max and Mad-Max recognizing DNA: Molecular bases of regulation by proto-oncogenic transcription factors
Cell(Cambridge,Mass.), 112, 2003
|
|
7ZJY
 
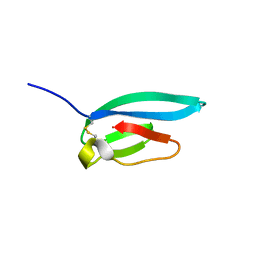 | | The NMR structure of the MAX67 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
7ZK0
 
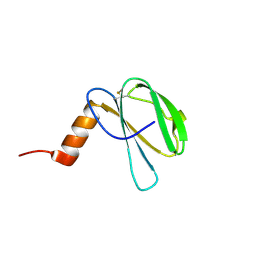 | | The NMR structure of the MAX60 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
7ZKD
 
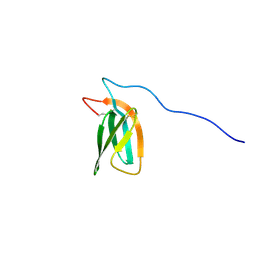 | | The NMR structure of the MAX47 effector from Magnaporthe Oryzae | | Descriptor: | MAX effector protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Pissarra, J, Raji, M, Cesari, S, Kroj, T, Gladieux, P, Roumestand, C, Barthe, P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
2MHW
 
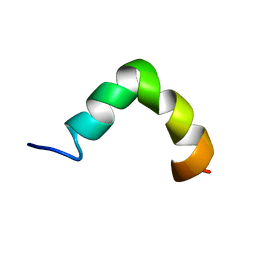 | | The solution NMR structure of maximin-4 in SDS micelles | | Descriptor: | Antimicrobial peptide | | Authors: | Toke, O, Banoczi, Z, Kiraly, P, Heinzmann, R, Burck, J, Ulrich, A.S, Hudecz, F. | | Deposit date: | 2013-12-05 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A kinked antimicrobial peptide from Bombina maxima. I. Three-dimensional structure determined by NMR in membrane-mimicking environments.
Eur.Biophys.J., 40, 2011
|
|
7SA1
 
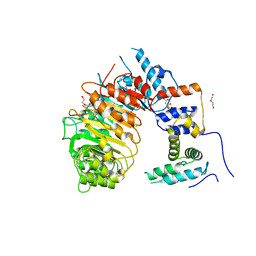 | | LRR-F-Box plant ubiquitin ligase | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, F-box/LRR-repeat MAX2 homolog, ... | | Authors: | Palayam, M, Shabek, N. | | Deposit date: | 2021-09-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | A conformational switch in the SCF-D3/MAX2 ubiquitin ligase facilitates strigolactone signalling.
Nat.Plants, 8, 2022
|
|
7NH9
 
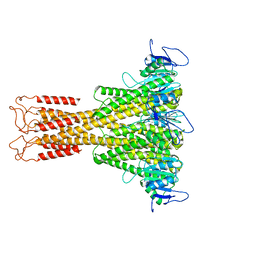 | |
2VO4
 
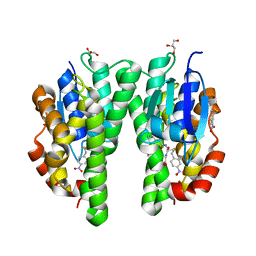 | | Glutathione transferase from Glycine max | | Descriptor: | 2,4-D INDUCIBLE GLUTATHIONE S-TRANSFERASE, 4-NITROPHENYL METHANETHIOL, GLYCEROL, ... | | Authors: | Axarli, I, Dhavala, P, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic and Functional Characterization of the Fluorodifen-Inducible Glutathione Transferase from Glycine Max Reveals an Active Site Topography Suited for Diphenylether Herbicides and a Novel L-Site.
J.Mol.Biol., 385, 2009
|
|
4TOP
 
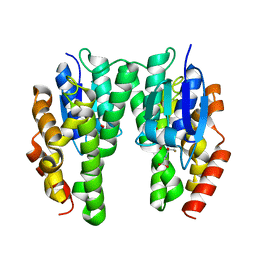 | | Glycine max glutathione transferase | | Descriptor: | 2,4-D inducible glutathione S-transferase, GLUTATHIONE | | Authors: | Axarli, I, Dhavala, P, Papageorgiou, A.C. | | Deposit date: | 2014-06-06 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Comparative analysis of the structural and functional features of two homologous tau class glutathione transferases from Glycine max
To Be Published
|
|
4CHS
 
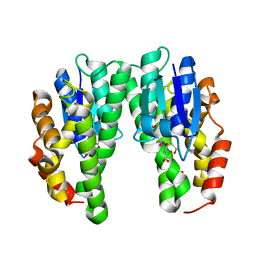 | | Crystal structure of a tau class glutathione transferase 10 from Glycine max | | Descriptor: | ACETONE, GLUTATHIONE S-TRANSFERASE, S-Hydroxy-Glutathione | | Authors: | Skopelitou, K, Muleta, A.W, Papageorgiou, A.C, Pavli, O, Flemetakis, E, Chronopoulou, E, Skaracis, G.N, Labrou, N.E. | | Deposit date: | 2013-12-04 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Catalytic features and crystal structure of a tau class glutathione transferase from Glycine max specifically upregulated in response to soybean mosaic virus infections.
Biochim. Biophys. Acta, 1854, 2015
|
|
2H5U
 
 | | Crystal structure of laccase from Cerrena maxima at 1.9A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lyashenko, A.V, Gabdoulkhakov, A.G, Zaitsev, V.N, Lamzin, V.S, Lindley, P.F, Bento, I, Betzel, C, Zhukhlistova, N.E, Zhukova, Y.N, Mikhailov, A.M. | | Deposit date: | 2006-05-27 | | Release date: | 2007-05-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Purification, crystallization and preliminary X-ray study of the fungal laccase from Cerrena maxima
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1F1F
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM ARTHROSPIRA MAXIMA | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Serag, A.A, Sawaya, M.R, Krogmann, D.W, Yeates, T.O. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
