5NP2
 
 | | Abl1 SH3 pTyr89/134 | | Descriptor: | Tyrosine-protein kinase ABL1 | | Authors: | Mero, B, Radnai, L, Gogl, G, Leveles, I, Buday, L. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the tyrosine phosphorylation-mediated inhibition of SH3 domain-ligand interactions.
J.Biol.Chem., 294, 2019
|
|
5O2P
 
 | | p130Cas SH3 domain PTP-PEST peptide chimera | | Descriptor: | Breast cancer anti-estrogen resistance 1,Tyrosine-protein phosphatase non-receptor type 12 | | Authors: | Hexnerova, R, Veverka, V. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CAS SH3 domain selectivity and regulation reveals new CAS interaction partners.
Sci Rep, 7, 2017
|
|
5OAZ
 
 | |
5OB0
 
 | |
5OB2
 
 | |
5OAV
 
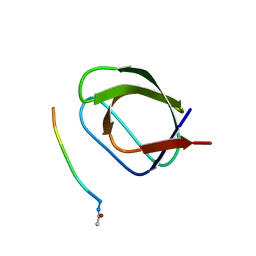 | |
5OB1
 
 | |
5O2Q
 
 | | p130Cas SH3 domain Vinculin peptide chimera | | Descriptor: | Breast cancer anti-estrogen resistance 1,Vinculin | | Authors: | Hexnerova, R, Veverka, V. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CAS SH3 domain selectivity and regulation reveals new CAS interaction partners.
Sci Rep, 7, 2017
|
|
5O2M
 
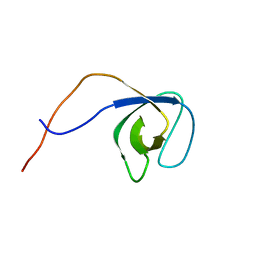 | | p130Cas SH3 domain | | Descriptor: | Breast cancer anti-estrogen resistance 1 | | Authors: | Hexnerova, R, Veverka, V. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CAS SH3 domain selectivity and regulation reveals new CAS interaction partners.
Sci Rep, 7, 2017
|
|
3SEM
 
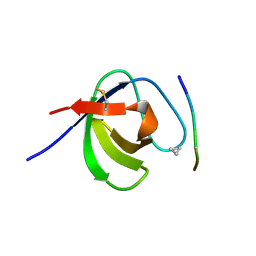 | | SEM5 SH3 DOMAIN COMPLEXED WITH PEPTOID INHIBITOR | | Descriptor: | SEX MUSCLE ABNORMAL PROTEIN 5, SH3 PEPTOID INHIBITOR | | Authors: | Nguyen, J.T, Turck, C.W, Cohen, F.E, Zuckermann, R.N, Lim, W.A. | | Deposit date: | 1998-11-02 | | Release date: | 1999-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors.
Science, 282, 1998
|
|
3THK
 
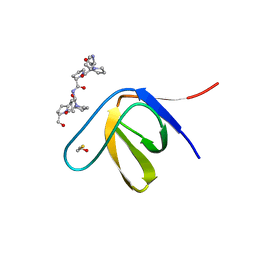 | | Structure of SH3 chimera with a type II ligand linked to the chain C-terminal | | Descriptor: | BETA-MERCAPTOETHANOL, Proline-rich peptide, SULFATE ION, ... | | Authors: | Gabdulkhakov, A.G, Gushchina, L.V, Nikulin, A.D, Nikonov, S.V, Filimonov, V.V. | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-Resolution Crystal Structure of Spectrin SH3 Domain Fused with a Proline-Rich Peptide.
J.Biomol.Struct.Dyn., 29, 2011
|
|
3UA6
 
 | |
3UA7
 
 | | Crystal Structure of the Human Fyn SH3 domain in complex with a peptide from the Hepatitis C virus NS5A-protein | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Martin-Garcia, J.M, Ruiz-Sanz, J, Luque, I, Camara-Artigas, A. | | Deposit date: | 2011-10-21 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The promiscuous binding of the Fyn SH3 domain to a peptide from the NS5A protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5QU8
 
 | |
5QU3
 
 | |
5QUA
 
 | | Crystal Structure of swapped human Nck SH3.1 domain, 1.5A, C2221 | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
5QU1
 
 | | Crystal Structure of the monomeric human Nck SH3.1 domain, triclinic, 1.08A | | Descriptor: | Cytoplasmic protein NCK1, SULFATE ION | | Authors: | Burger, D, Ruf, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
5QU5
 
 | | Domain Swap in the first SH3 domain of human Nck1 | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Burger, D, Ruf, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
5QU2
 
 | | Crystal Structure of human Nck SH3.1 in complex with peptide PPPVPNPDY | | Descriptor: | ACE-PRO-PRO-PRO-VAL-PRO-ASN-PRO-ASP-TYR-NH2, Cytoplasmic protein NCK1, SULFATE ION | | Authors: | Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
5QU7
 
 | |
5QU4
 
 | |
5QU6
 
 | | Crystal Structure of swapped human Nck SH3.1 domain, 1.8A, triclinic | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
1CKB
 
 | |
1CKA
 
 | |
1CSK
 
 | |
