5D4C
 
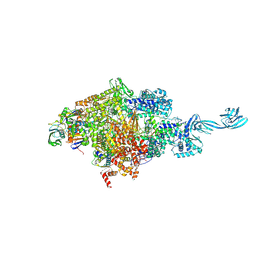 | |
1L0O
 
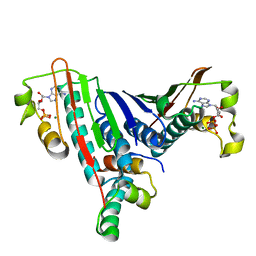 | | Crystal Structure of the Bacillus stearothermophilus Anti-Sigma Factor SpoIIAB with the Sporulation Sigma Factor SigmaF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anti-sigma F factor, MAGNESIUM ION, ... | | Authors: | Campbell, E.A, Masuda, S, Sun, J.L, Muzzin, O, Olson, C.A, Wang, S, Darst, S.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the Bacillus stearothermophilus anti-sigma factor SpoIIAB with the sporulation sigma factor sigmaF.
Cell(Cambridge,Mass.), 108, 2002
|
|
1L9U
 
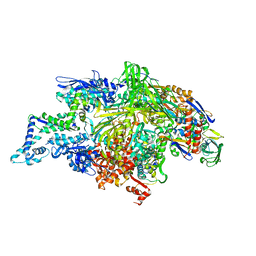 | | THERMUS AQUATICUS RNA POLYMERASE HOLOENZYME AT 4 A RESOLUTION | | Descriptor: | MAGNESIUM ION, RNA POLYMERASE, ALPHA SUBUNIT, ... | | Authors: | Murakami, K.S, Masuda, S, Darst, S.A. | | Deposit date: | 2002-03-26 | | Release date: | 2002-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution.
Science, 296, 2002
|
|
1L9Z
 
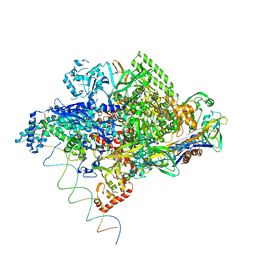 | | Thermus aquaticus RNA Polymerase Holoenzyme/Fork-Junction Promoter DNA Complex at 6.5 A Resolution | | Descriptor: | MAGNESIUM ION, RNA POLYMERASE, ALPHA SUBUNIT, ... | | Authors: | Murakami, K.S, Masuda, S, Campbell, E.A, Muzzin, O, Darst, S.A. | | Deposit date: | 2002-03-27 | | Release date: | 2002-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex.
Science, 296, 2002
|
|
8X6G
 
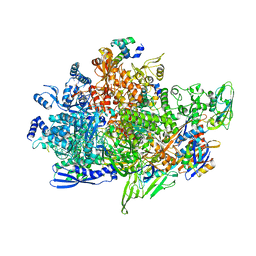 | | Cryo-EM structure of Staphylococcus aureus sigB-dependent RNAP-promoter open complex | | Descriptor: | DNA (70-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yuan, L, Xu, L, Liu, Q, Feng, Y. | | Deposit date: | 2023-11-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of promoter recognition by Staphylococcus aureus RNA polymerase.
Nat Commun, 15, 2024
|
|
8X6F
 
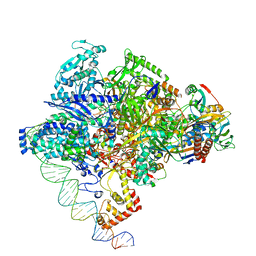 | | Cryo-EM structure of Staphylococcus aureus sigA-dependent RNAP-promoter open complex | | Descriptor: | DNA (71-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yuan, L, Xu, L, Liu, Q, Feng, Y. | | Deposit date: | 2023-11-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of promoter recognition by Staphylococcus aureus RNA polymerase.
Nat Commun, 15, 2024
|
|
8W8N
 
 | |
8W8P
 
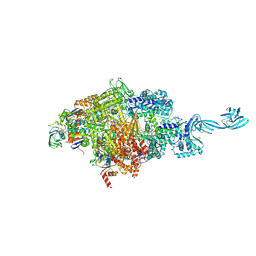 | |
8W8O
 
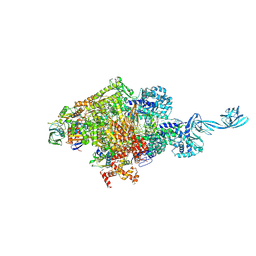 | |
8Y6U
 
 | |
8U3B
 
 | | Cryo-EM structure of E. coli NarL-transcription activation complex at 3.2A | | Descriptor: | DNA (69-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Kompaniiets, D, Wang, D. | | Deposit date: | 2023-09-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis for transcription activation by the nitrate-responsive regulator NarL.
Nucleic Acids Res., 52, 2024
|
|
8TOE
 
 | |
8TO8
 
 | | Escherichia coli RNA polymerase unwinding intermediate (I1b) at the lambda PR promoter | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
8TO1
 
 | | Escherichia coli RNA polymerase unwinding intermediate (I1a) at the lambda PR promoter | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
8TO6
 
 | | Escherichia coli RNA polymerase unwinding intermediate (I1d) at the lambda PR promoter | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
8TOM
 
 | |
6P1K
 
 | | Cryo-EM structure of Escherichia coli sigma70 bound RNAP polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-05-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
6P18
 
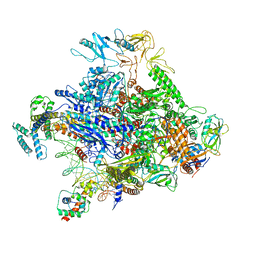 | | Q21 transcription antitermination complex: loading complex | | Descriptor: | DNA (67-MER) fragment carrying phage-21 pR' promoter and pause element, nontemplate strand, template strand, ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of Q-dependent antitermination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6PMJ
 
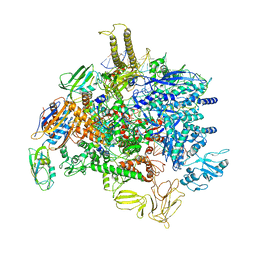 | | Sigm28-transcription initiation complex with specific promoter at the state 2 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
6P70
 
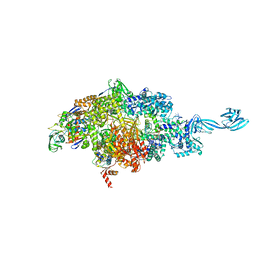 | | X-ray crystal structure of bacterial RNA polymerase and pyrBI promoter complex | | Descriptor: | DNA (5'-D(*TP*AP*TP*AP*AP*TP*CP*GP*AP*TP*CP*TP*TP*TP*GP*CP*CP*GP*GP*G)-3'), DNA (5'-D(P*TP*CP*CP*CP*GP*GP*CP*AP*AP*AP*TP*TP*GP*TP*CP*CP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shin, Y, Murakami, K.S. | | Deposit date: | 2019-06-04 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Structural basis of reiterative transcription from the pyrG and pyrBI promoters by bacterial RNA polymerase.
Nucleic Acids Res., 48, 2020
|
|
6PB4
 
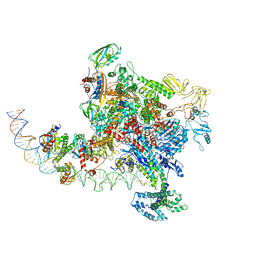 | |
6P71
 
 | |
6PB5
 
 | |
4MEY
 
 | | Crystal structure of Escherichia coli RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Feng, Y, Zhang, Y, Arnold, E, Ebright, R.H. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.948 Å) | | Cite: | Transcription inhibition by the depsipeptide antibiotic salinamide A.
Elife, 3, 2014
|
|
8GZH
 
 | |
