3ANV
 
 | | Crystal structure of D-serine dehydratase from chicken kidney (2,3-DAP complex) | | Descriptor: | 3-amino-D-alanine, CHLORIDE ION, D-serine dehydratase, ... | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2010-09-09 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney
J.Biol.Chem., 286, 2011
|
|
3AWN
 
 | | Crystal structure of D-serine dehydratase from chicken kidney (EDTA treated) | | Descriptor: | D-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney.
J.Biol.Chem., 286, 2011
|
|
3AWO
 
 | | Crystal structure of D-serine dehydratase in complex with D-serine from chicken kidney (EDTA-treated) | | Descriptor: | D-SERINE, D-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney.
J.Biol.Chem., 286, 2011
|
|
3B8T
 
 | | Crystal structure of Escherichia coli alaine racemase mutant P219A | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3B8U
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221A | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3B8V
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221K | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3B8W
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221P | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3CO8
 
 | | Crystal structure of alanine racemase from Oenococcus oeni | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Palani, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of alanine racemase from Oenococcus oeni with bound pyridoxal 5'-phosphate.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3CPG
 
 | |
3E5P
 
 | | Crystal structure of alanine racemase from E.faecalis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alanine racemase, NONAETHYLENE GLYCOL, ... | | Authors: | Hwang, K.Y, Priyadarshi, A, Lee, E.H, Sung, M.W. | | Deposit date: | 2008-08-14 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the alanine racemase from Enterococcus faecalis.
Biochim.Biophys.Acta, 1794, 2009
|
|
3E6E
 
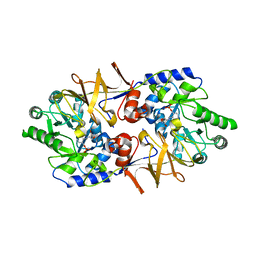 | | Crystal structure of Alanine racemase from E.faecalis complex with cycloserine | | Descriptor: | Alanine racemase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Hwang, K.Y, Priyadarshi, A, Lee, E.H, Sung, M.W. | | Deposit date: | 2008-08-15 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the alanine racemase from Enterococcus faecalis.
Biochim.Biophys.Acta, 1794, 2009
|
|
3GWQ
 
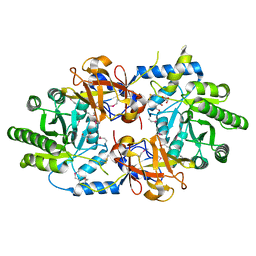 | |
3HA1
 
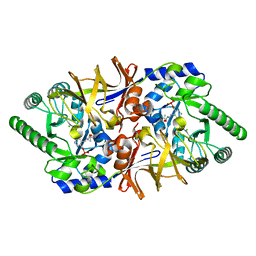 | | Alanine racemase from Bacillus Anthracis (Ames) | | Descriptor: | ACETATE ION, Alanine racemase, CHLORIDE ION | | Authors: | Counago, R.M, Davlieva, M, Strych, U, Hill, R.E, Krause, K.L. | | Deposit date: | 2009-04-30 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural characterization of alanine racemase from Bacillus anthracis (Ames).
Bmc Struct.Biol., 9, 2009
|
|
3HUR
 
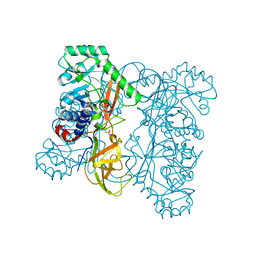 | | Crystal structure of alanine racemase from Oenococcus oeni | | Descriptor: | Alanine racemase, SULFATE ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Hu, S, Romero, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-15 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of alanine racemase from Oenococcus oeni
To be Published
|
|
3KW3
 
 | |
3LLX
 
 | |
3OO2
 
 | | 2.37 Angstrom resolution crystal structure of an alanine racemase (alr) from Staphylococcus aureus subsp. aureus COL | | Descriptor: | Alanine racemase 1, BETA-MERCAPTOETHANOL, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Winsor, J, Dubrovska, I, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-30 | | Release date: | 2010-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | 2.37 Angstrom resolution crystal structure of an alanine racemase (alr) from Staphylococcus aureus subsp. aureus COL
TO BE PUBLISHED
|
|
3R79
 
 | |
3S46
 
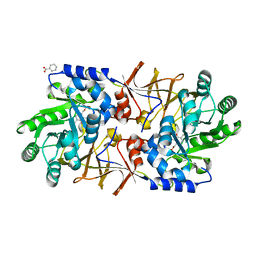 | | The crystal structure of alanine racemase from streptococcus pneumoniae | | Descriptor: | Alanine racemase, BENZOIC ACID | | Authors: | Im, H, Sharpe, M.L, Strych, U, Davlieva, M, Krause, K.L. | | Deposit date: | 2011-05-18 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of alanine racemase from Streptococcus pneumoniae, a target for structure-based drug design.
BMC MICROBIOL., 11, 2011
|
|
3SY1
 
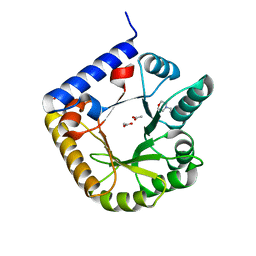 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR70 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, UPF0001 protein yggS | | Authors: | Vorobiev, S, Su, M, Nivon, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Maglaqui, M, Baker, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.465 Å) | | Cite: | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR70
To be Published
|
|
3UW6
 
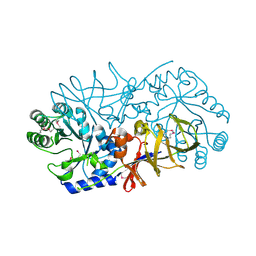 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR120 | | Descriptor: | Alanine racemase | | Authors: | Seetharaman, J, Lew, S, Nivon, L, Baker, D, Bjelic, S, Ciccosanti, C, Sahdev, S, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-11-30 | | Release date: | 2012-02-08 | | Last modified: | 2022-03-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of enone-binding proteins with catalytic activity for the Morita-Baylis-Hillman reaction.
Acs Chem.Biol., 8, 2013
|
|
3WQC
 
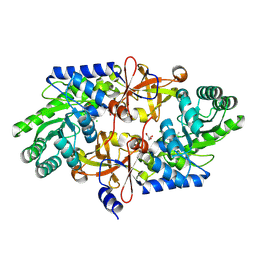 | | D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23 | | Descriptor: | CHLORIDE ION, D-threo-3-hydroxyaspartate dehydratase, GLYCEROL, ... | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-01-25 | | Release date: | 2015-01-28 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WQD
 
 | | D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23 complexed with D-erythro-3-hydroxyaspartate | | Descriptor: | (3S)-3-hydroxy-D-aspartic acid, D-threo-3-hydroxyaspartate dehydratase, MAGNESIUM ION, ... | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-01-25 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WQE
 
 | | D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23 complexed with D-allothreonine | | Descriptor: | D-allothreonine, D-threo-3-hydroxyaspartate dehydratase, MAGNESIUM ION, ... | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-01-25 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WQF
 
 | |
