Functional annotation example
[Functional Annotation of] 1uan
1uan (TT1542 from Thermus thermophilus HB8) is a structural genomics (SG) target solved as part of the Protein 3000 project in Japan [1]. At the time the structure was solved, there were no PDB entries with significant sequence homology, however conservation and geometry of surface-exposed residues enabled the identification of a putative active site near theHPDDGEloop located near the N-terminus. The authors then looked for similar ligand binding sites in the eF-site database. (The procedure has since been implemented in the eF-seek server.) Based on similarity to several enzymes (mycothiol S-conjugate amidase and GlcNAc-Ins deacetylase, from Mycobacterium tuberculosis and GlcNAc-PI de-N-acetylases from several eukaryotes), 1uan was proposed to be a hydrolase acting on a disaccharide substrate.
Since the 1uan structure was published, the crystal structures of several GlcNAc-Ins deacetylases from Mycobacterium tuberculosis have been solved (e.g., 1q7t, 1q74 ). These structures can be identified by Sequence Navigator and Structure Navigator and support the location of the proposed active site near the conserved loop (in 1q74 the loop has the sequenceHPDDES) as well as the overall function of 1uan: GlcNAc-Ins deacetylases remove an acetyl group from a disacharide in the mycothiol synthetic pathway. Currently, the closest match to 1uan in the PDB is 2ixd, a putative deacetylase from Bacillus cereus that was co-crystallized with zinc and acetate[1].
[Usage]
Here we will demonstrate how Structure Navigator can be used to locate homologs of 1uan.

In the PDB Code box type1uan. In the Chain box type eitherAorB(1uanA is 99% homologous to 1uanB). Next press theSearchbutton.
Message window should appear when the job is over:

Choosing "Job manager" button, "Job manager" should appear:

Clicking on the "Done" link, will send you to the following page (Only the first few lines are shown):

We see that the first hit is 1uan itself (chain B, if you entered 1uan A as the query). The first non-self hit is 2ixdA. Next you can see the entries 1q74 and 1q7t. Clicking on the 2ixdA link, will send you to the Mine summary page:

Going back one page, and clicking on the alignment link for 2ixdA will give the structure alignment:

Can you find the conserved loop?
If you have installed jV on your system, you can view the superposition in 3D by clicking the jV version 3 link.
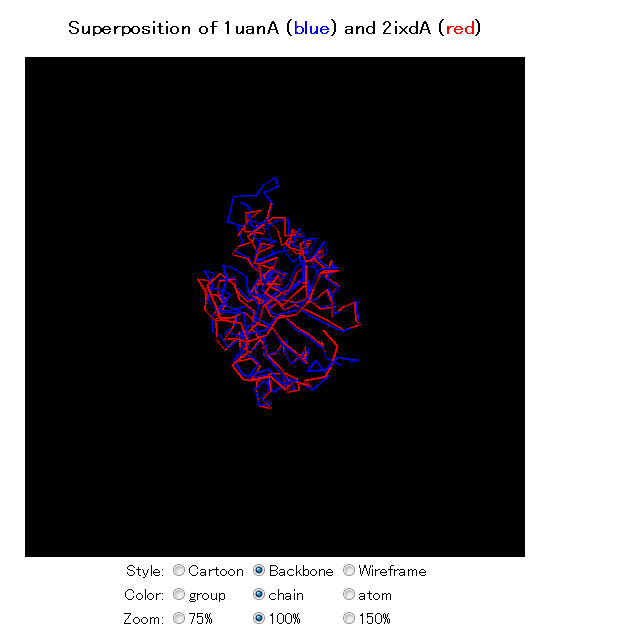
Can you find the active site based on the position of the conserved loop?
[References]
1. Handa N, Terada T, Kamewari Y, Hamana H, Tame JR, Park SY, Kinoshita K, Ota M, Nakamura H, Kuramitsu S, Shirouzu M, Yokoyama S. Crystal structure of the conserved protein TT1542 from Thermus thermophilus HB8. Protein Sci 2003;12(8):1621-1632.






