[English] 日本語
 Yorodumi
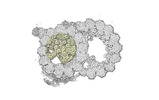
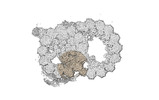
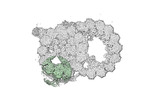
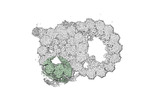
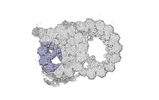
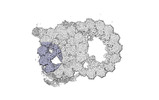
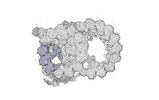
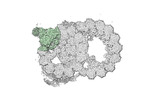
Yorodumi- PDB-8snb: atomic model of sea urchin sperm doublet microtubule (48-nm perio... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8snb | ||||||
|---|---|---|---|---|---|---|---|
| Title | atomic model of sea urchin sperm doublet microtubule (48-nm periodicity) | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  STRUCTURAL PROTEIN / STRUCTURAL PROTEIN /  sperm / doublet microtubule / DMT / microtubule inner protein / sperm / doublet microtubule / DMT / microtubule inner protein /  cilia / cilia /  flagella / FAP / CFAP / MIP / MAP / flagella / FAP / CFAP / MIP / MAP /  cytoskeleton / tublin / cytoskeleton / tublin /  sea urchin / TEX / sea urchin / TEX /  Tektin / SAXO / ODF / SPATA / RIB Tektin / SAXO / ODF / SPATA / RIB | ||||||
| Function / homology |  Function and homology information Function and homology informationcilium-dependent cell motility / regulation of cilium beat frequency involved in ciliary motility / : / cilium movement involved in cell motility /  axoneme assembly / axonemal microtubule / cilium organization / axoneme assembly / axonemal microtubule / cilium organization /  nucleoside-diphosphate kinase / UTP biosynthetic process / CTP biosynthetic process ...cilium-dependent cell motility / regulation of cilium beat frequency involved in ciliary motility / : / cilium movement involved in cell motility / nucleoside-diphosphate kinase / UTP biosynthetic process / CTP biosynthetic process ...cilium-dependent cell motility / regulation of cilium beat frequency involved in ciliary motility / : / cilium movement involved in cell motility /  axoneme assembly / axonemal microtubule / cilium organization / axoneme assembly / axonemal microtubule / cilium organization /  nucleoside-diphosphate kinase / UTP biosynthetic process / CTP biosynthetic process / nucleoside-diphosphate kinase / UTP biosynthetic process / CTP biosynthetic process /  motile cilium / positive regulation of cell motility / GTP biosynthetic process / motile cilium / positive regulation of cell motility / GTP biosynthetic process /  nucleoside diphosphate kinase activity / ciliary base / regulation of neuron projection development / nucleoside diphosphate kinase activity / ciliary base / regulation of neuron projection development /  axoneme / alpha-tubulin binding / mitotic cytokinesis / axoneme / alpha-tubulin binding / mitotic cytokinesis /  cilium assembly / sperm flagellum / cilium assembly / sperm flagellum /  phosphatase binding / microtubule-based process / phosphatase binding / microtubule-based process /  Hsp70 protein binding / mitotic spindle organization / acrosomal vesicle / ciliary basal body / Hsp70 protein binding / mitotic spindle organization / acrosomal vesicle / ciliary basal body /  Hsp90 protein binding / structural constituent of cytoskeleton / Hsp90 protein binding / structural constituent of cytoskeleton /  mitotic spindle / microtubule cytoskeleton organization / microtubule cytoskeleton / mitotic cell cycle / protein-folding chaperone binding / mitotic spindle / microtubule cytoskeleton organization / microtubule cytoskeleton / mitotic cell cycle / protein-folding chaperone binding /  microtubule binding / microtubule binding /  spermatogenesis / vesicle / spermatogenesis / vesicle /  microtubule / microtubule /  cytoskeleton / cytoskeleton /  calmodulin binding / calmodulin binding /  hydrolase activity / neuron projection / hydrolase activity / neuron projection /  phosphorylation / phosphorylation /  GTPase activity / GTPase activity /  centrosome / centrosome /  calcium ion binding / GTP binding / positive regulation of transcription by RNA polymerase II / calcium ion binding / GTP binding / positive regulation of transcription by RNA polymerase II /  ATP binding / ATP binding /  nucleus / nucleus /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Strongylocentrotus purpuratus (purple sea urchin) Strongylocentrotus purpuratus (purple sea urchin) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.3 Å cryo EM / Resolution: 3.3 Å | ||||||
 Authors Authors | Zeng, J. / Zhang, R. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Structural specializations of the sperm tail. Authors: Miguel Ricardo Leung / Jianwei Zeng / Xiangli Wang / Marc C Roelofs / Wei Huang / Riccardo Zenezini Chiozzi / Johannes F Hevler / Albert J R Heck / Susan K Dutcher / Alan Brown / Rui Zhang / ...Authors: Miguel Ricardo Leung / Jianwei Zeng / Xiangli Wang / Marc C Roelofs / Wei Huang / Riccardo Zenezini Chiozzi / Johannes F Hevler / Albert J R Heck / Susan K Dutcher / Alan Brown / Rui Zhang / Tzviya Zeev-Ben-Mordehai /   Abstract: Sperm motility is crucial to reproductive success in sexually reproducing organisms. Impaired sperm movement causes male infertility, which is increasing globally. Sperm are powered by a microtubule- ...Sperm motility is crucial to reproductive success in sexually reproducing organisms. Impaired sperm movement causes male infertility, which is increasing globally. Sperm are powered by a microtubule-based molecular machine-the axoneme-but it is unclear how axonemal microtubules are ornamented to support motility in diverse fertilization environments. Here, we present high-resolution structures of native axonemal doublet microtubules (DMTs) from sea urchin and bovine sperm, representing external and internal fertilizers. We identify >60 proteins decorating sperm DMTs; at least 15 are sperm associated and 16 are linked to infertility. By comparing DMTs across species and cell types, we define core microtubule inner proteins (MIPs) and analyze evolution of the tektin bundle. We identify conserved axonemal microtubule-associated proteins (MAPs) with unique tubulin-binding modes. Additionally, we identify a testis-specific serine/threonine kinase that links DMTs to outer dense fibers in mammalian sperm. Our study provides structural foundations for understanding sperm evolution, motility, and dysfunction at a molecular level. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8snb.cif.gz 8snb.cif.gz | 26.5 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8snb.ent.gz pdb8snb.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  8snb.json.gz 8snb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/sn/8snb https://data.pdbj.org/pub/pdb/validation_reports/sn/8snb ftp://data.pdbj.org/pub/pdb/validation_reports/sn/8snb ftp://data.pdbj.org/pub/pdb/validation_reports/sn/8snb | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  40619MC  8otzC  8ou0C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
+Protein , 41 types, 395 molecules 1A1B1P1Q1T1U1V1W1Y1d1f1g1i1j9M9N9O1l1m2A2B2C2D2G2O3A3B3C3D3E...
-Coiled-coil domain-containing protein ... , 2 types, 13 molecules 1E1F1G1H1K1L1M1v1w1x1y1z2a
| #2: Protein | Mass: 49945.527 Da / Num. of mol.: 4 / Source method: isolated from a natural source Source: (natural)   Strongylocentrotus purpuratus (purple sea urchin) Strongylocentrotus purpuratus (purple sea urchin)References: UniProt: A0A7M7RBY5 #3: Protein | Mass: 79691.844 Da / Num. of mol.: 9 / Source method: isolated from a natural source Source: (natural)   Strongylocentrotus purpuratus (purple sea urchin) Strongylocentrotus purpuratus (purple sea urchin)References: UniProt: A0A7M7PGM9 |
|---|
-Outer dense fiber protein ... , 2 types, 17 molecules 1a1b5E5F5G5H5I5J5K5L5M5N5O5A5B9Y9Z
| #7: Protein | Mass: 27280.830 Da / Num. of mol.: 13 / Source method: isolated from a natural source Source: (natural)   Strongylocentrotus purpuratus (purple sea urchin) Strongylocentrotus purpuratus (purple sea urchin)References: UniProt: A0A7M7RI41 #30: Protein | Mass: 30624.738 Da / Num. of mol.: 4 / Source method: isolated from a natural source Source: (natural)   Strongylocentrotus purpuratus (purple sea urchin) Strongylocentrotus purpuratus (purple sea urchin)References: UniProt: A0A7M7RDG4 |
|---|
-Meiosis-specific nuclear structural protein ... , 2 types, 6 molecules 1o1p1q1r2R2S
| #12: Protein | Mass: 17323.562 Da / Num. of mol.: 4 / Source method: isolated from a natural source Source: (natural)   Strongylocentrotus purpuratus (purple sea urchin) Strongylocentrotus purpuratus (purple sea urchin)References: UniProt: A0A7M7NFX5 #17: Protein | Mass: 61705.172 Da / Num. of mol.: 2 / Source method: isolated from a natural source Source: (natural)   Strongylocentrotus purpuratus (purple sea urchin) Strongylocentrotus purpuratus (purple sea urchin)References: UniProt: A0A7M7REB1 |
|---|
-Cilia- and flagella-associated protein ... , 6 types, 19 molecules 2J2K2L2V2W3W3X3Y3Z4A4B4C4N4O4P4Q4T4U4V
| #15: Protein | Mass: 15883.667 Da / Num. of mol.: 3 / Source method: isolated from a natural source Source: (natural)   Strongylocentrotus purpuratus (purple sea urchin) Strongylocentrotus purpuratus (purple sea urchin)References: UniProt: A0A7M7NA77 #18: Protein | Mass: 32480.740 Da / Num. of mol.: 2 / Source method: isolated from a natural source Source: (natural)   Strongylocentrotus purpuratus (purple sea urchin) Strongylocentrotus purpuratus (purple sea urchin)References: UniProt: A0A7M7RAY9 #23: Protein | Mass: 64116.262 Da / Num. of mol.: 4 / Source method: isolated from a natural source Source: (natural)   Strongylocentrotus purpuratus (purple sea urchin) Strongylocentrotus purpuratus (purple sea urchin)References: UniProt: A0A7M7NFL8 #24: Protein | Mass: 70455.148 Da / Num. of mol.: 3 / Source method: isolated from a natural source Source: (natural)   Strongylocentrotus purpuratus (purple sea urchin) Strongylocentrotus purpuratus (purple sea urchin)References: UniProt: A0A7M7NV05 #27: Protein | Mass: 27690.299 Da / Num. of mol.: 4 / Source method: isolated from a natural source Source: (natural)   Strongylocentrotus purpuratus (purple sea urchin) Strongylocentrotus purpuratus (purple sea urchin)References: UniProt: A0A7M7TG06 #28: Protein | Mass: 27701.848 Da / Num. of mol.: 3 / Source method: isolated from a natural source Source: (natural)   Strongylocentrotus purpuratus (purple sea urchin) Strongylocentrotus purpuratus (purple sea urchin)References: UniProt: A0A7M7GHD6 |
|---|
-Trichohyalin-plectin-homology domain-containing ... , 2 types, 4 molecules 3N3O4F4G
| #21: Protein | Mass: 65783.406 Da / Num. of mol.: 2 / Source method: isolated from a natural source Source: (natural)   Strongylocentrotus purpuratus (purple sea urchin) Strongylocentrotus purpuratus (purple sea urchin)References: UniProt: A0A7M7REW2 #25: Protein | Mass: 62040.836 Da / Num. of mol.: 2 / Source method: isolated from a natural source Source: (natural)   Strongylocentrotus purpuratus (purple sea urchin) Strongylocentrotus purpuratus (purple sea urchin)References: UniProt: A0A7M7RHS4 |
|---|
-Non-polymers , 3 types, 446 molecules 




| #56: Chemical | ChemComp-GTP /  Guanosine triphosphate Guanosine triphosphate#57: Chemical | ChemComp-MG / #58: Chemical | ChemComp-GDP /  Guanosine diphosphate Guanosine diphosphate |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: FILAMENT / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: sea urchin sperm doublet microtubule / Type: ORGANELLE OR CELLULAR COMPONENT / Entity ID: #1-#55 / Source: NATURAL |
|---|---|
| Source (natural) | Organism:   Strongylocentrotus purpuratus (purple sea urchin) Strongylocentrotus purpuratus (purple sea urchin) |
| Buffer solution | pH: 7.4 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2500 nm / Nominal defocus min: 500 nm Bright-field microscopy / Nominal defocus max: 2500 nm / Nominal defocus min: 500 nm |
| Image recording | Electron dose: 34 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
- Processing
Processing
| Software | Name: UCSF ChimeraX / Version: 1.5/v9 / Classification: model building / URL: https://www.rbvi.ucsf.edu/chimerax/ / Os: macOS / Type: package |
|---|---|
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION |
3D reconstruction | Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 127673 / Symmetry type: POINT |
 Movie
Movie Controller
Controller




















 PDBj
PDBj





