[English] 日本語
 Yorodumi
Yorodumi- EMDB-17188: bovine sperm endpiece singlet microtubules (one tubulin dimer and... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | bovine sperm endpiece singlet microtubules (one tubulin dimer and associated microtubule inner proteins) | |||||||||
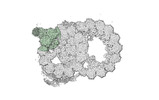
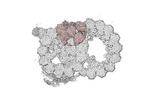
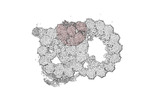
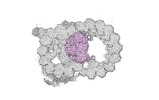
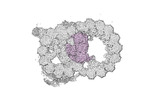
 Map data Map data | cryo-EM map obtained after symmetry expansion showing two tubulin dimers along a protofilament from bovine sperm endpiece microtubules | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  microtubule / microtubule inner protein / microtubule / microtubule inner protein /  sperm / sperm /  cilia / cilia /  CYTOSOLIC PROTEIN CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationMicrotubule-dependent trafficking of connexons from Golgi to the plasma membrane /  Cilium Assembly / Cilium Assembly /  Intraflagellar transport / Carboxyterminal post-translational modifications of tubulin / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / Resolution of Sister Chromatid Cohesion / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / COPI-dependent Golgi-to-ER retrograde traffic ...Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Intraflagellar transport / Carboxyterminal post-translational modifications of tubulin / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / Resolution of Sister Chromatid Cohesion / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / COPI-dependent Golgi-to-ER retrograde traffic ...Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane /  Cilium Assembly / Cilium Assembly /  Intraflagellar transport / Carboxyterminal post-translational modifications of tubulin / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / Resolution of Sister Chromatid Cohesion / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / COPI-dependent Golgi-to-ER retrograde traffic / RHO GTPases activate IQGAPs / COPI-independent Golgi-to-ER retrograde traffic / COPI-mediated anterograde transport / RHO GTPases Activate Formins / cold acclimation / MHC class II antigen presentation / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / Intraflagellar transport / Carboxyterminal post-translational modifications of tubulin / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / Resolution of Sister Chromatid Cohesion / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / COPI-dependent Golgi-to-ER retrograde traffic / RHO GTPases activate IQGAPs / COPI-independent Golgi-to-ER retrograde traffic / COPI-mediated anterograde transport / RHO GTPases Activate Formins / cold acclimation / MHC class II antigen presentation / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand /  axoneme assembly / axonemal microtubule / Aggrephagy / The role of GTSE1 in G2/M progression after G2 checkpoint / Separation of Sister Chromatids / Loss of Nlp from mitotic centrosomes / Recruitment of mitotic centrosome proteins and complexes / Loss of proteins required for interphase microtubule organization from the centrosome / Anchoring of the basal body to the plasma membrane / AURKA Activation by TPX2 / Recruitment of NuMA to mitotic centrosomes / axoneme assembly / axonemal microtubule / Aggrephagy / The role of GTSE1 in G2/M progression after G2 checkpoint / Separation of Sister Chromatids / Loss of Nlp from mitotic centrosomes / Recruitment of mitotic centrosome proteins and complexes / Loss of proteins required for interphase microtubule organization from the centrosome / Anchoring of the basal body to the plasma membrane / AURKA Activation by TPX2 / Recruitment of NuMA to mitotic centrosomes /  Regulation of PLK1 Activity at G2/M Transition / positive regulation of cilium assembly / Hedgehog 'off' state / Neutrophil degranulation / cellular response to cold / ciliary base / Regulation of PLK1 Activity at G2/M Transition / positive regulation of cilium assembly / Hedgehog 'off' state / Neutrophil degranulation / cellular response to cold / ciliary base /  intercellular bridge / sperm flagellum / intercellular bridge / sperm flagellum /  centriole / acrosomal vesicle / ciliary basal body / centriole / acrosomal vesicle / ciliary basal body /  Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / structural constituent of cytoskeleton / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / structural constituent of cytoskeleton /  mitotic spindle / microtubule cytoskeleton organization / calcium-dependent protein binding / mitotic spindle / microtubule cytoskeleton organization / calcium-dependent protein binding /  double-stranded RNA binding / mitotic cell cycle / double-stranded RNA binding / mitotic cell cycle /  microtubule binding / microtubule binding /  microtubule / protein stabilization / microtubule / protein stabilization /  cytoskeleton / cytoskeleton /  hydrolase activity / hydrolase activity /  GTPase activity / GTP binding / GTPase activity / GTP binding /  metal ion binding / metal ion binding /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Bos taurus (cattle) / Bos taurus (cattle) /   cattle (cattle) cattle (cattle) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.5 Å cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Leung MR / Zeev-Ben-Mordehai T | |||||||||
| Funding support |  Netherlands, 1 items Netherlands, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Structural specializations of the sperm tail. Authors: Miguel Ricardo Leung / Jianwei Zeng / Xiangli Wang / Marc C Roelofs / Wei Huang / Riccardo Zenezini Chiozzi / Johannes F Hevler / Albert J R Heck / Susan K Dutcher / Alan Brown / Rui Zhang / ...Authors: Miguel Ricardo Leung / Jianwei Zeng / Xiangli Wang / Marc C Roelofs / Wei Huang / Riccardo Zenezini Chiozzi / Johannes F Hevler / Albert J R Heck / Susan K Dutcher / Alan Brown / Rui Zhang / Tzviya Zeev-Ben-Mordehai /   Abstract: Sperm motility is crucial to reproductive success in sexually reproducing organisms. Impaired sperm movement causes male infertility, which is increasing globally. Sperm are powered by a microtubule- ...Sperm motility is crucial to reproductive success in sexually reproducing organisms. Impaired sperm movement causes male infertility, which is increasing globally. Sperm are powered by a microtubule-based molecular machine-the axoneme-but it is unclear how axonemal microtubules are ornamented to support motility in diverse fertilization environments. Here, we present high-resolution structures of native axonemal doublet microtubules (DMTs) from sea urchin and bovine sperm, representing external and internal fertilizers. We identify >60 proteins decorating sperm DMTs; at least 15 are sperm associated and 16 are linked to infertility. By comparing DMTs across species and cell types, we define core microtubule inner proteins (MIPs) and analyze evolution of the tektin bundle. We identify conserved axonemal microtubule-associated proteins (MAPs) with unique tubulin-binding modes. Additionally, we identify a testis-specific serine/threonine kinase that links DMTs to outer dense fibers in mammalian sperm. Our study provides structural foundations for understanding sperm evolution, motility, and dysfunction at a molecular level. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17188.map.gz emd_17188.map.gz | 4.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17188-v30.xml emd-17188-v30.xml emd-17188.xml emd-17188.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_17188.png emd_17188.png | 109.5 KB | ||
| Others |  emd_17188_half_map_1.map.gz emd_17188_half_map_1.map.gz emd_17188_half_map_2.map.gz emd_17188_half_map_2.map.gz | 97.5 MB 97.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17188 http://ftp.pdbj.org/pub/emdb/structures/EMD-17188 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17188 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17188 | HTTPS FTP |
-Related structure data
| Related structure data |  8ou0MC  8otzC  8snbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17188.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17188.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
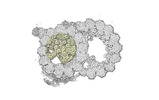
| Annotation | cryo-EM map obtained after symmetry expansion showing two tubulin dimers along a protofilament from bovine sperm endpiece microtubules | ||||||||||||||||||||||||||||||||||||
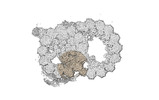
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.359 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half-map1
| File | emd_17188_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
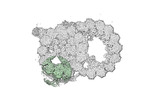
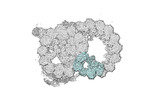
| Annotation | half-map1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half-map2
| File | emd_17188_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
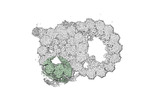
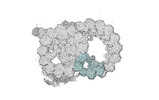
| Annotation | half-map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : tubulin heterodimer with associated microtubule inner proteins SP...
| Entire | Name: tubulin heterodimer with associated microtubule inner proteins SPACA9 and SAXO1 |
|---|---|
| Components |
|
-Supramolecule #1: tubulin heterodimer with associated microtubule inner proteins SP...
| Supramolecule | Name: tubulin heterodimer with associated microtubule inner proteins SPACA9 and SAXO1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Bos taurus (cattle) Bos taurus (cattle) |
-Macromolecule #1: Tubulin beta-4B chain
| Macromolecule | Name: Tubulin beta-4B chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   cattle (cattle) cattle (cattle) |
| Molecular weight | Theoretical: 49.877824 KDa |
| Sequence | String: MREIVHLQAG QCGNQIGAKF WEVISDEHGI DPTGTYHGDS DLQLERINVY YNEATGGKYV PRAVLVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKEAESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS ...String: MREIVHLQAG QCGNQIGAKF WEVISDEHGI DPTGTYHGDS DLQLERINVY YNEATGGKYV PRAVLVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKEAESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS VVPSPKVSDT VVEPYNATLS VHQLVENTDE TYCIDNEALY DICFRTLKLT TPTYGDLNHL VSATMSGVTT CL RFPGQLN ADLRKLAVNM VPFPRLHFFM PGFAPLTSRG SQQYRALTVP ELTQQMFDAK NMMAACDPRH GRYLTVAAVF RGR MSMKEV DEQMLNVQNK NSSYFVEWIP NNVKTAVCDI PPRGLKMSAT FIGNSTAIQE LFKRISEQFT AMFRRKAFLH WYTG EGMDE MEFTEAESNM NDLVSEYQQY QDATAEEEGE FEEEAEEEVA |
-Macromolecule #2: Stabilizer of axonemal microtubules 1
| Macromolecule | Name: Stabilizer of axonemal microtubules 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   cattle (cattle) cattle (cattle) |
| Molecular weight | Theoretical: 54.67207 KDa |
| Sequence | String: MAPTKGKCVC ELCSCGRHHC PHLPTKIYDK TEKPCLLSEY TENYPVYHSY LPRESFKPKM DYQRACTPME GLTTSRRDFG PHKVLPVKI HQPNPFVPSE ENMDLQTTYK QDYNPYPLCR VDPFKPRDSK YPCGDKMESL PTYKADYLPW NQPRRELLRP P HHYRPAST ...String: MAPTKGKCVC ELCSCGRHHC PHLPTKIYDK TEKPCLLSEY TENYPVYHSY LPRESFKPKM DYQRACTPME GLTTSRRDFG PHKVLPVKI HQPNPFVPSE ENMDLQTTYK QDYNPYPLCR VDPFKPRDSK YPCGDKMESL PTYKADYLPW NQPRRELLRP P HHYRPAST KFDSRTTQQD DYSMKGLVNT RSCKPPAVPK LCNVPLEDLT NYKMSYVAHP LEKRFVHESE KFRPCEIPFE SL TTHKESY RGLMGEPAKS LKPPARPYGL DTPFSNTTEF RDKYQAWPTP QVFSKPPSMY VPPEEKMDLL TTVQTHYTYP KGA PAESCR PALSVKKGGR FEGSTTTKED YKQWASTRTE PAKPIPQLNL PTEPLDCLTT ARAHYVPHLP MMTKSCKPVW SGPQ GNIPV EGQTTYTISF TPKEMSRCLA SYPEPPGYIF EEIDALGHRI YRPVSQTGSR RSSRFSVGDS ENPNQQELTV SA |
-Macromolecule #3: Sperm acrosome associated 9
| Macromolecule | Name: Sperm acrosome associated 9 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   cattle (cattle) cattle (cattle) |
| Molecular weight | Theoretical: 25.174801 KDa |
| Sequence | String: MMNEVKESLR SVEQKYKIFQ QQQFTFIGAL EHCRENAHDK IRPISSIGQV QSYMEHHCSN STDRRILLMF LDICSELSKL CQHFEALHA GTPVTNNLLE KCKTLVSQSN DLSSLRAKYP HDVVNHLSCD EARNHYGGVV SLIPIILDLM KEWVAHSEKL P RKALQQVS ...String: MMNEVKESLR SVEQKYKIFQ QQQFTFIGAL EHCRENAHDK IRPISSIGQV QSYMEHHCSN STDRRILLMF LDICSELSKL CQHFEALHA GTPVTNNLLE KCKTLVSQSN DLSSLRAKYP HDVVNHLSCD EARNHYGGVV SLIPIILDLM KEWVAHSEKL P RKALQQVS EPQAATRATA HAPQASGTQP QLRKQNCGQL IQNIPKPGGK DQGSSKPPWR PPGGKL |
-Macromolecule #4: Tubulin alpha-3 chain
| Macromolecule | Name: Tubulin alpha-3 chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO EC number:  Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:   cattle (cattle) cattle (cattle) |
| Molecular weight | Theoretical: 49.978344 KDa |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVVDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIVDLVL DRIRKLADLC TGLQGFLIFH SFGGGTGSGF ASLLMERLSV D YGKKSKLE ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVVDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIVDLVL DRIRKLADLC TGLQGFLIFH SFGGGTGSGF ASLLMERLSV D YGKKSKLE FAIYPAPQVS TAVVEPYNSI LTTHTTLEHS DCAFMVDNEA IYDICRRNLD IERPTYTNLN RLIGQIVSSI TA SLRFDGA LNVDLTEFQT NLVPYPRIHF PLATYAPVIS AEKAYHEQLS VAEITNACFE PANQMVKCDP RHGKYMACCM LYR GDVVPK DVNAAIATIK TKRTIQFVDW CPTGFKVGIN YQPPTVVPGG DLAKVQRAVC MLSNTTAIAE AWARLDHKLD LMYA KRAFV HWYVGEGMEE GEFSEAREDL AALEKDYEEV GVDSVEAEAE EGEEY |
-Macromolecule #5: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Macromolecule #6: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 1 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information |  ChemComp-GTP: |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.3 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.1) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.1) |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 240292 |
 Movie
Movie Controller
Controller









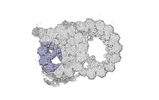
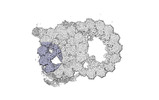
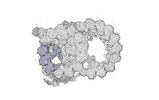
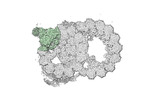
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































