[English] 日本語
 Yorodumi
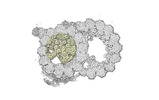
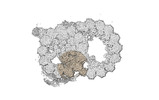
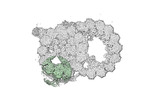
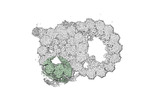
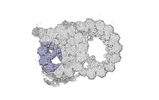
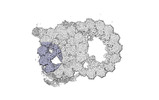
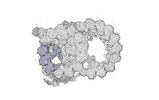
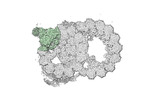
Yorodumi- PDB-8ou0: bovine sperm endpiece singlet microtubules (one tubulin dimer and... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8ou0 | ||||||
|---|---|---|---|---|---|---|---|
| Title | bovine sperm endpiece singlet microtubules (one tubulin dimer and associated microtubule inner proteins) | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  CYTOSOLIC PROTEIN / CYTOSOLIC PROTEIN /  microtubule / microtubule inner protein / microtubule / microtubule inner protein /  sperm / sperm /  cilia cilia | ||||||
| Function / homology |  Function and homology information Function and homology informationMicrotubule-dependent trafficking of connexons from Golgi to the plasma membrane /  Cilium Assembly / Cilium Assembly /  Intraflagellar transport / Carboxyterminal post-translational modifications of tubulin / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / Resolution of Sister Chromatid Cohesion / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / COPI-dependent Golgi-to-ER retrograde traffic ...Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Intraflagellar transport / Carboxyterminal post-translational modifications of tubulin / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / Resolution of Sister Chromatid Cohesion / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / COPI-dependent Golgi-to-ER retrograde traffic ...Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane /  Cilium Assembly / Cilium Assembly /  Intraflagellar transport / Carboxyterminal post-translational modifications of tubulin / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / Resolution of Sister Chromatid Cohesion / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / COPI-dependent Golgi-to-ER retrograde traffic / RHO GTPases activate IQGAPs / COPI-independent Golgi-to-ER retrograde traffic / COPI-mediated anterograde transport / RHO GTPases Activate Formins / cold acclimation / MHC class II antigen presentation / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / Intraflagellar transport / Carboxyterminal post-translational modifications of tubulin / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / Resolution of Sister Chromatid Cohesion / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / COPI-dependent Golgi-to-ER retrograde traffic / RHO GTPases activate IQGAPs / COPI-independent Golgi-to-ER retrograde traffic / COPI-mediated anterograde transport / RHO GTPases Activate Formins / cold acclimation / MHC class II antigen presentation / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand /  axoneme assembly / axonemal microtubule / Aggrephagy / The role of GTSE1 in G2/M progression after G2 checkpoint / Separation of Sister Chromatids / Loss of Nlp from mitotic centrosomes / Recruitment of mitotic centrosome proteins and complexes / Loss of proteins required for interphase microtubule organization from the centrosome / Anchoring of the basal body to the plasma membrane / AURKA Activation by TPX2 / Recruitment of NuMA to mitotic centrosomes / axoneme assembly / axonemal microtubule / Aggrephagy / The role of GTSE1 in G2/M progression after G2 checkpoint / Separation of Sister Chromatids / Loss of Nlp from mitotic centrosomes / Recruitment of mitotic centrosome proteins and complexes / Loss of proteins required for interphase microtubule organization from the centrosome / Anchoring of the basal body to the plasma membrane / AURKA Activation by TPX2 / Recruitment of NuMA to mitotic centrosomes /  Regulation of PLK1 Activity at G2/M Transition / positive regulation of cilium assembly / Hedgehog 'off' state / Neutrophil degranulation / cellular response to cold / ciliary base / Regulation of PLK1 Activity at G2/M Transition / positive regulation of cilium assembly / Hedgehog 'off' state / Neutrophil degranulation / cellular response to cold / ciliary base /  intercellular bridge / sperm flagellum / intercellular bridge / sperm flagellum /  centriole / acrosomal vesicle / ciliary basal body / centriole / acrosomal vesicle / ciliary basal body /  Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / structural constituent of cytoskeleton / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / structural constituent of cytoskeleton /  mitotic spindle / microtubule cytoskeleton organization / calcium-dependent protein binding / mitotic spindle / microtubule cytoskeleton organization / calcium-dependent protein binding /  double-stranded RNA binding / mitotic cell cycle / double-stranded RNA binding / mitotic cell cycle /  microtubule binding / microtubule binding /  microtubule / protein stabilization / microtubule / protein stabilization /  cytoskeleton / cytoskeleton /  hydrolase activity / hydrolase activity /  GTPase activity / GTP binding / GTPase activity / GTP binding /  metal ion binding / metal ion binding /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Bos taurus (cattle) Bos taurus (cattle) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.5 Å cryo EM / Resolution: 3.5 Å | ||||||
 Authors Authors | Leung, M.R. / Zeev-Ben-Mordehai, T. | ||||||
| Funding support |  Netherlands, 1items Netherlands, 1items
| ||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Structural specializations of the sperm tail. Authors: Miguel Ricardo Leung / Jianwei Zeng / Xiangli Wang / Marc C Roelofs / Wei Huang / Riccardo Zenezini Chiozzi / Johannes F Hevler / Albert J R Heck / Susan K Dutcher / Alan Brown / Rui Zhang / ...Authors: Miguel Ricardo Leung / Jianwei Zeng / Xiangli Wang / Marc C Roelofs / Wei Huang / Riccardo Zenezini Chiozzi / Johannes F Hevler / Albert J R Heck / Susan K Dutcher / Alan Brown / Rui Zhang / Tzviya Zeev-Ben-Mordehai /   Abstract: Sperm motility is crucial to reproductive success in sexually reproducing organisms. Impaired sperm movement causes male infertility, which is increasing globally. Sperm are powered by a microtubule- ...Sperm motility is crucial to reproductive success in sexually reproducing organisms. Impaired sperm movement causes male infertility, which is increasing globally. Sperm are powered by a microtubule-based molecular machine-the axoneme-but it is unclear how axonemal microtubules are ornamented to support motility in diverse fertilization environments. Here, we present high-resolution structures of native axonemal doublet microtubules (DMTs) from sea urchin and bovine sperm, representing external and internal fertilizers. We identify >60 proteins decorating sperm DMTs; at least 15 are sperm associated and 16 are linked to infertility. By comparing DMTs across species and cell types, we define core microtubule inner proteins (MIPs) and analyze evolution of the tektin bundle. We identify conserved axonemal microtubule-associated proteins (MAPs) with unique tubulin-binding modes. Additionally, we identify a testis-specific serine/threonine kinase that links DMTs to outer dense fibers in mammalian sperm. Our study provides structural foundations for understanding sperm evolution, motility, and dysfunction at a molecular level. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8ou0.cif.gz 8ou0.cif.gz | 198.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8ou0.ent.gz pdb8ou0.ent.gz | 153.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8ou0.json.gz 8ou0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ou/8ou0 https://data.pdbj.org/pub/pdb/validation_reports/ou/8ou0 ftp://data.pdbj.org/pub/pdb/validation_reports/ou/8ou0 ftp://data.pdbj.org/pub/pdb/validation_reports/ou/8ou0 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  17188MC  8otzC  8snbC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 4 types, 4 molecules BDCA
| #1: Protein |  / Tubulin beta-2C chain / Tubulin beta-2C chainMass: 49877.824 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Bos taurus (cattle) / References: UniProt: Q3MHM5 Bos taurus (cattle) / References: UniProt: Q3MHM5 |
|---|---|
| #2: Protein | Mass: 54672.070 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Bos taurus (cattle) / References: UniProt: A0A3S5ZPV0 Bos taurus (cattle) / References: UniProt: A0A3S5ZPV0 |
| #3: Protein | Mass: 25174.801 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Bos taurus (cattle) / References: UniProt: A0A3Q1MYU9 Bos taurus (cattle) / References: UniProt: A0A3Q1MYU9 |
| #4: Protein | Mass: 49978.344 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Bos taurus (cattle) Bos taurus (cattle)References: UniProt: Q32KN8,  Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement |
-Non-polymers , 3 types, 3 molecules 




| #5: Chemical | ChemComp-GDP /  Guanosine diphosphate Guanosine diphosphate |
|---|---|
| #6: Chemical | ChemComp-GTP /  Guanosine triphosphate Guanosine triphosphate |
| #7: Chemical | ChemComp-MG / |
-Details
| Has ligand of interest | N |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: FILAMENT / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: tubulin heterodimer with associated microtubule inner proteins SPACA9 and SAXO1 Type: COMPLEX / Entity ID: #1-#4 / Source: NATURAL |
|---|---|
| Source (natural) | Organism:   Bos taurus (cattle) Bos taurus (cattle) |
| Buffer solution | pH: 7.3 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
| Specimen support | Grid material: COPPER / Grid mesh size: 200 divisions/in. / Grid type: Quantifoil R2/1 |
Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TALOS ARCTICA |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2500 nm / Nominal defocus min: 500 nm / Alignment procedure: COMA FREE Bright-field microscopy / Nominal defocus max: 2500 nm / Nominal defocus min: 500 nm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN |
| Image recording | Electron dose: 50 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.19.2_4158: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
3D reconstruction | Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 240292 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller




















 PDBj
PDBj






