-Search query
-Search result
Showing 1 - 50 of 262 items for (author: jensen & g & j)
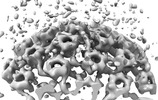
EMDB-18482: 
Herpes simplex virus 1 capsid (WT) vertices in perinuclear NEC-coated vesicles determined in situ

EMDB-18484: 
Herpes simplex virus 1 nuclear egress complex (WT) determined in situ from perinuclear vesicles

EMDB-17974: 
Pseudorabies virus cytosolic C-capsid (US3 KO) vertices determined in situ

EMDB-17975: 
Pseudorabies virus primary enveloped (perinuclear) C-capsid (US3 KO) vertices determined in situ

EMDB-17976: 
Pseudorabies nuclear C-capsids (US3 KO) vertices determined in situ

EMDB-18473: 
Subtomogram average of pseudorabies virus nuclear egress complex helical form (UL31/34) determined in situ

EMDB-18474: 
Subtomogram average of pseudorabies virus nuclear egress complex (UL31/34) determined in situ

EMDB-18479: 
Pseudorabies virus cytosolic C-capsid (WT) vertices determined in situ

EMDB-18480: 
Pseudorabies virus nuclear C-capsid (WT) vertices determined in situ

EMDB-18481: 
Herpes simplex virus 1 cytosolic C-capsid (WT) vertices determined in situ
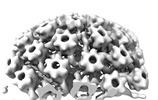
EMDB-18483: 
Herpes simplex virus 1 nuclear C-capsid (WT) vertices determined in situ

EMDB-19895: 
Structure of IgE HMM5 bound to FceRIa cryo-EM class 8

PDB-9eq3: 
Structure of IgE HMM5 bound to FceRIa cryo-EM class 8

EMDB-19896: 
Structure of IgE HMM5 bound to FceRIa cryo-EM class 5

PDB-9eq4: 
Structure of IgE HMM5 bound to FceRIa cryo-EM class 5

EMDB-28957: 
Cryo-EM structure of the Agrobacterium T-pilus

PDB-8fai: 
Cryo-EM structure of the Agrobacterium T-pilus

EMDB-16321: 
Structure of the human nuclear cap-binding complex bound to NCBP3(560-620) and cap-analogue m7GpppG

PDB-8by6: 
Structure of the human nuclear cap-binding complex bound to NCBP3(560-620) and cap-analogue m7GpppG

EMDB-17763: 
Structure of the human nuclear cap-binding complex bound to ARS2[147-871] and m7GTP

EMDB-17784: 
Structure of the human nuclear cap-binding complex bound to PHAX and m7G-capped RNA

PDB-8pmp: 
Structure of the human nuclear cap-binding complex bound to ARS2[147-871] and m7GTP

PDB-8pnt: 
Structure of the human nuclear cap-binding complex bound to PHAX and m7G-capped RNA

EMDB-28281: 
Subtomogram average of the T4SS of Coxiella Burnetii at pH 4.75
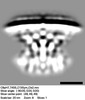
EMDB-28282: 
Subtomogram average of T4SS of Coxiella burnetii at pH 7
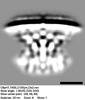
EMDB-28283: 
Subtomogram average of T4SS of Coxiella burnetii at pH7 with an inner membrane mask

EMDB-16416: 
HB3VAR03 apo headstructure (PfEMP1 A) complexed with EPCR

PDB-8c44: 
HB3VAR03 apo headstructure (PfEMP1 A) complexed with EPCR

EMDB-16415: 
HB3VAR03 apo headstructure (PfEMP1 A)

PDB-8c3y: 
HB3VAR03 apo headstructure (PfEMP1 A)

EMDB-29916: 
Subtomogram average of the AnaS GV shell

EMDB-29921: 
Subtomogram average of the native Ana GV shell

PDB-8gbs: 
Integrative model of the native Ana GV shell

EMDB-26571: 
Subtomogram average of a non-treated cellulose fiber (related to Figure 7A of primary citation)

EMDB-26572: 
Subtomogram average of a bapta cellulose fiber (related to Figure 7B of primary citation)

EMDB-26573: 
Subtomogram average of a pectate lyase cellulose fiber (related to Figure 7C of primary citation)

EMDB-29922: 
Cryo-tomogram of the native Ana GV

EMDB-29923: 
Cryo-tomogram of the Halo GV (c-vac)

EMDB-29924: 
Cryo-tomogram of Halo GV (p-vac)

EMDB-29925: 
Cryo-tomogram of the Mega GVs

EMDB-16377: 
Focused map for structure of IgE bound to the ectodomain of FceRIa

EMDB-16378: 
Structure of IgE bound to the ectodomain of FceRIa

PDB-8c1b: 
Focused map for structure of IgE bound to the ectodomain of FceRIa

PDB-8c1c: 
Structure of IgE bound to the ectodomain of FceRIa

EMDB-25214: 
Cryo-EM structure of Drosophila Integrator cleavage module (IntS4-IntS9-IntS11) in complex with IP6

PDB-7sn8: 
Cryo-EM structure of Drosophila Integrator cleavage module (IntS4-IntS9-IntS11) in complex with IP6

EMDB-14874: 
Cryo-EM Structure of Human Transferrin Receptor 1 bound to DNA Aptamer

PDB-7zqs: 
Cryo-EM Structure of Human Transferrin Receptor 1 bound to DNA Aptamer
EMDB-27654: 
A subtomogram average of H. neapolitanus Rubisco within alpha-carboxysomes

EMDB-14777: 
Cryo-EM structure of archaic chaperone-usher Csu pilus of Acinetobacter baumannii
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
