[English] 日本語
 Yorodumi
Yorodumi- EMDB-16321: Structure of the human nuclear cap-binding complex bound to NCBP3... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human nuclear cap-binding complex bound to NCBP3(560-620) and cap-analogue m7GpppG | |||||||||
 Map data Map data | primary map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nuclear cap-binding complex / NCBP3 / Pol II transcript metabolism /  RNA BINDING PROTEIN RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsnRNA export from nucleus / nuclear cap binding complex / mRNA metabolic process / RNA cap binding complex / histone mRNA metabolic process / positive regulation of mRNA 3'-end processing / positive regulation of RNA export from nucleus / cap-dependent translational initiation / Processing of Intronless Pre-mRNAs /  snRNA binding ...snRNA export from nucleus / nuclear cap binding complex / mRNA metabolic process / RNA cap binding complex / histone mRNA metabolic process / positive regulation of mRNA 3'-end processing / positive regulation of RNA export from nucleus / cap-dependent translational initiation / Processing of Intronless Pre-mRNAs / snRNA binding ...snRNA export from nucleus / nuclear cap binding complex / mRNA metabolic process / RNA cap binding complex / histone mRNA metabolic process / positive regulation of mRNA 3'-end processing / positive regulation of RNA export from nucleus / cap-dependent translational initiation / Processing of Intronless Pre-mRNAs /  snRNA binding / positive regulation of RNA binding / snRNA binding / positive regulation of RNA binding /  RNA cap binding / alternative mRNA splicing, via spliceosome / SLBP independent Processing of Histone Pre-mRNAs / SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs / RNA cap binding / alternative mRNA splicing, via spliceosome / SLBP independent Processing of Histone Pre-mRNAs / SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs /  regulation of mRNA processing / primary miRNA processing / regulatory ncRNA-mediated post-transcriptional gene silencing / miRNA-mediated post-transcriptional gene silencing / : / Transport of the SLBP independent Mature mRNA / Transport of the SLBP Dependant Mature mRNA / RNA 7-methylguanosine cap binding / mRNA 3'-end processing / mRNA 3'-end processing / Transport of Mature mRNA Derived from an Intronless Transcript / mRNA cis splicing, via spliceosome / positive regulation of mRNA splicing, via spliceosome / RNA catabolic process / Transport of Mature mRNA derived from an Intron-Containing Transcript / Abortive elongation of HIV-1 transcript in the absence of Tat / FGFR2 alternative splicing / regulation of translational initiation / RNA Polymerase II Transcription Termination / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / Signaling by FGFR2 IIIa TM / spliceosomal complex assembly / Formation of the Early Elongation Complex / Formation of the HIV-1 Early Elongation Complex / mRNA Capping / mRNA Splicing - Minor Pathway / 7-methylguanosine mRNA capping / Processing of Capped Intron-Containing Pre-mRNA / RNA polymerase II transcribes snRNA genes / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Formation of HIV-1 elongation complex containing HIV-1 Tat / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Formation of HIV elongation complex in the absence of HIV Tat / mRNA export from nucleus / Formation of RNA Pol II elongation complex / RNA Polymerase II Pre-transcription Events / mRNA Splicing - Major Pathway / regulation of mRNA processing / primary miRNA processing / regulatory ncRNA-mediated post-transcriptional gene silencing / miRNA-mediated post-transcriptional gene silencing / : / Transport of the SLBP independent Mature mRNA / Transport of the SLBP Dependant Mature mRNA / RNA 7-methylguanosine cap binding / mRNA 3'-end processing / mRNA 3'-end processing / Transport of Mature mRNA Derived from an Intronless Transcript / mRNA cis splicing, via spliceosome / positive regulation of mRNA splicing, via spliceosome / RNA catabolic process / Transport of Mature mRNA derived from an Intron-Containing Transcript / Abortive elongation of HIV-1 transcript in the absence of Tat / FGFR2 alternative splicing / regulation of translational initiation / RNA Polymerase II Transcription Termination / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / Signaling by FGFR2 IIIa TM / spliceosomal complex assembly / Formation of the Early Elongation Complex / Formation of the HIV-1 Early Elongation Complex / mRNA Capping / mRNA Splicing - Minor Pathway / 7-methylguanosine mRNA capping / Processing of Capped Intron-Containing Pre-mRNA / RNA polymerase II transcribes snRNA genes / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Formation of HIV-1 elongation complex containing HIV-1 Tat / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Formation of HIV elongation complex in the absence of HIV Tat / mRNA export from nucleus / Formation of RNA Pol II elongation complex / RNA Polymerase II Pre-transcription Events / mRNA Splicing - Major Pathway /  RNA splicing / positive regulation of transcription elongation by RNA polymerase II / mRNA transcription by RNA polymerase II / RNA splicing / positive regulation of transcription elongation by RNA polymerase II / mRNA transcription by RNA polymerase II /  mRNA splicing, via spliceosome / Regulation of expression of SLITs and ROBOs / mRNA splicing, via spliceosome / Regulation of expression of SLITs and ROBOs /  snRNP Assembly / positive regulation of cell growth / defense response to virus / molecular adaptor activity / nuclear speck / snRNP Assembly / positive regulation of cell growth / defense response to virus / molecular adaptor activity / nuclear speck /  ribonucleoprotein complex / ribonucleoprotein complex /  mRNA binding / mRNA binding /  mitochondrion / mitochondrion /  DNA binding / DNA binding /  RNA binding / RNA binding /  nucleoplasm / nucleoplasm /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
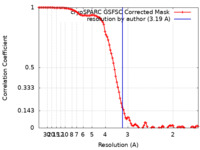
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.19 Å cryo EM / Resolution: 3.19 Å | |||||||||
 Authors Authors | Dubiez E / Pellegrini E / Foucher AE / Cusack S / Kadlec J | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2024 Journal: Cell Rep / Year: 2024Title: Structural basis for competitive binding of productive and degradative co-transcriptional effectors to the nuclear cap-binding complex. Authors: Etienne Dubiez / Erika Pellegrini / Maja Finderup Brask / William Garland / Anne-Emmanuelle Foucher / Karine Huard / Torben Heick Jensen / Stephen Cusack / Jan Kadlec /   Abstract: The nuclear cap-binding complex (CBC) coordinates co-transcriptional maturation, transport, or degradation of nascent RNA polymerase II (Pol II) transcripts. CBC with its partner ARS2 forms mutually ...The nuclear cap-binding complex (CBC) coordinates co-transcriptional maturation, transport, or degradation of nascent RNA polymerase II (Pol II) transcripts. CBC with its partner ARS2 forms mutually exclusive complexes with diverse "effectors" that promote either productive or destructive outcomes. Combining AlphaFold predictions with structural and biochemical validation, we show how effectors NCBP3, NELF-E, ARS2, PHAX, and ZC3H18 form competing binary complexes with CBC and how PHAX, NCBP3, ZC3H18, and other effectors compete for binding to ARS2. In ternary CBC-ARS2 complexes with PHAX, NCBP3, or ZC3H18, ARS2 is responsible for the initial effector recruitment but inhibits their direct binding to the CBC. We show that in vivo ZC3H18 binding to both CBC and ARS2 is required for nuclear RNA degradation. We propose that recruitment of PHAX to CBC-ARS2 can lead, with appropriate cues, to competitive displacement of ARS2 and ZC3H18 from the CBC, thus promoting a productive rather than a degradative RNA fate. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16321.map.gz emd_16321.map.gz | 51.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16321-v30.xml emd-16321-v30.xml emd-16321.xml emd-16321.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16321_fsc.xml emd_16321_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_16321.png emd_16321.png | 73.4 KB | ||
| Masks |  emd_16321_msk_1.map emd_16321_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16321.cif.gz emd-16321.cif.gz | 6.2 KB | ||
| Others |  emd_16321_additional_1.map.gz emd_16321_additional_1.map.gz emd_16321_half_map_1.map.gz emd_16321_half_map_1.map.gz emd_16321_half_map_2.map.gz emd_16321_half_map_2.map.gz | 3.5 MB 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16321 http://ftp.pdbj.org/pub/emdb/structures/EMD-16321 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16321 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16321 | HTTPS FTP |
-Related structure data
| Related structure data |  8by6MC  8pmpC  8pntC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16321.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16321.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | primary map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.827 Å | ||||||||||||||||||||||||||||||||||||
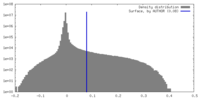
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16321_msk_1.map emd_16321_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
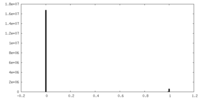
| Density Histograms |
-Additional map: locally filtered and sharpened map (-100 A2)
| File | emd_16321_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | locally filtered and sharpened map (-100 A2) | ||||||||||||
| Projections & Slices |
| ||||||||||||
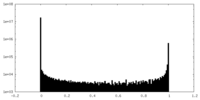
| Density Histograms |
-Half map: half map A
| File | emd_16321_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
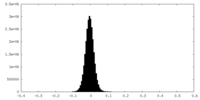
| Density Histograms |
-Half map: half map B
| File | emd_16321_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of CBP80, CBP20 and NCBP3 with bound m7GpppG
| Entire | Name: Ternary complex of CBP80, CBP20 and NCBP3 with bound m7GpppG |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of CBP80, CBP20 and NCBP3 with bound m7GpppG
| Supramolecule | Name: Ternary complex of CBP80, CBP20 and NCBP3 with bound m7GpppG type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Nuclear cap-binding protein subunit 1
| Macromolecule | Name: Nuclear cap-binding protein subunit 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 89.809984 KDa |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKTSDANETE DHLESLICKV GEKSACSLES NLEGLAGVLE ADLPNYKSKI LRLLCTVARL LPEKLTIYTT LVGLLNARNY NFGGEFVEA MIRQLKESLK ANNYNEAVYL VRFLSDLVNC HVIAAPSMVA MFENFVSVTQ EEDVPQVRRD WYVYAFLSSL P WVGKELYE ...String: MKTSDANETE DHLESLICKV GEKSACSLES NLEGLAGVLE ADLPNYKSKI LRLLCTVARL LPEKLTIYTT LVGLLNARNY NFGGEFVEA MIRQLKESLK ANNYNEAVYL VRFLSDLVNC HVIAAPSMVA MFENFVSVTQ EEDVPQVRRD WYVYAFLSSL P WVGKELYE KKDAEMDRIF ANTESYLKRR QKTHVPMLQV WTADKPHPQE EYLDCLWAQI QKLKKDRWQE RHILRPYLAF DS ILCEALQ HNLPPFTPPP HTEDSVYPMP RVIFRMFDYT DDPEGPVMPG SHSVERFVIE ENLHCIIKSH WKERKTCAAQ LVS YPGKNK IPLNYHIVEV IFAELFQLPA PPHIDVMYTT LLIELCKLQP GSLPQVLAQA TEMLYMRLDT MNTTCVDRFI NWFS HHLSN FQFRWSWEDW SDCLSQDPES PKPKFVREVL EKCMRLSYHQ RILDIVPPTF SALCPVNPTC IYKYGDESSN SLPGH SVAL CLAVAFKSKA TNDEIFSILK DVPNPNQDDD DDEGFSFNPL KIEVFVQTLL HLAAKSFSHS FSALAKFHEV FKTLAE SDE GKLHVLRVMF EVWRNHPQMI AVLVDKMIRT QIVDCAAVAN WIFSSELSRD FTRLFVWEIL HSTIRKMNKH VLKIQKE LE EAKEKLARQH KRRSDDDDRS SDRKDGVLEE QIERLQEKVE SAQSEQKNLF LVIFQRFIMI LTEHLVRCET DGTSVLTP W YKNCIERLQQ IFLQHHQIIQ QYMVTLENLL FTAELDPHIL AVFQQFCALQ A UniProtKB: Nuclear cap-binding protein subunit 1 |
-Macromolecule #2: Nuclear cap-binding protein subunit 2
| Macromolecule | Name: Nuclear cap-binding protein subunit 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.15626 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: GAMSGGLLKA LRSDSYVELS QYRDQHFRGD NEEQEKLLKK SCTLYVGNLS FYTTEEQIYE LFSKSGDIKK IIMGLDKMKK TACGFCFVE YYSRADAENA MRYINGTRLD DRIIRTDWDA GFKEGRQYGR GRSGGQVRDE YRQDYDAGRG GYGKLAQNQ UniProtKB: Nuclear cap-binding protein subunit 2 |
-Macromolecule #3: Nuclear cap-binding protein subunit 3
| Macromolecule | Name: Nuclear cap-binding protein subunit 3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 6.734328 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: KKVDHRAPGA EEDDSELQRA WGALIKEKEQ SRQKKSRLDN LPSLQIEVSR ESSSGSEAES UniProtKB: Nuclear cap-binding protein subunit 3 |
-Macromolecule #4: 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE
| Macromolecule | Name: 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE / type: ligand / ID: 4 / Number of copies: 1 / Formula: GTG |
|---|---|
| Molecular weight | Theoretical: 803.44 Da |
| Chemical component information |  ChemComp-GTG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7000000000000001 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7000000000000001 µm |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)