-Search query
-Search result
Showing 1 - 50 of 74 items for (author: snijder & j)
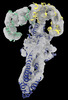
EMDB-18149: 
GBP1 bound by 14-3-3sigma
Method: single particle / : Pfleiderer MM, Liu X, Fisch D, Anastasakou E, Frickel EM, Galej WP

PDB-8q4l: 
GBP1 bound by 14-3-3sigma
Method: single particle / : Pfleiderer MM, Liu X, Fisch D, Anastasakou E, Frickel EM, Galej WP

EMDB-16882: 
Human Coronavirus HKU1 spike glycoprotein
Method: single particle / : Drulyte I, Hurdiss DL

EMDB-17076: 
Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (closed state)
Method: single particle / : Drulyte I, Hurdiss DL

EMDB-17077: 
Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (1-up state)
Method: single particle / : Drulyte I, Hurdiss DL

EMDB-17078: 
Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (3-up state)
Method: single particle / : Drulyte I, Hurdiss DL

EMDB-17079: 
Human Coronavirus HKU1 W89A spike glycoprotein incubated with an alpha2,8-linked 9-O-acetylated disialoside (closed state)
Method: single particle / : Drulyte I, Hurdiss DL

EMDB-17080: 
Local refinement of the Human Coronavirus HKU1 spike glycoprotein
Method: single particle / : Drulyte I, Hurdiss DL

EMDB-17081: 
Local refinement of the Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (closed state)
Method: single particle / : Drulyte I, Hurdiss DL

EMDB-17082: 
Local refinement of the Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (3-up state)
Method: single particle / : Drulyte I, Hurdiss DL

EMDB-17083: 
Local refinement of the Human Coronavirus HKU1 W89A spike glycoprotein incubated with an alpha2,8-linked 9-O-acetylated disialoside (closed state)
Method: single particle / : Drulyte I, Hurdiss DL

PDB-8ohn: 
Human Coronavirus HKU1 spike glycoprotein
Method: single particle / : Pronker MF, Hurdiss DL

PDB-8opm: 
Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (closed state)
Method: single particle / : Pronker MF, Creutznacher R, Hurdiss DL

PDB-8opn: 
Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (1-up state)
Method: single particle / : Pronker MF, Creutznacher R, Hurdiss DL

PDB-8opo: 
Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (3-up state)
Method: single particle / : Pronker MF, Creutznacher R, Hurdiss DL

EMDB-16144: 
Structure of the SARS-CoV-2 spike glycoprotein in complex with the macrocyclic peptide S1B3inL1
Method: single particle / : Hurdiss DL, Drulyte I

PDB-8bon: 
Structure of the SARS-CoV-2 spike glycoprotein in complex with the macrocyclic peptide S1B3inL1
Method: single particle / : Hurdiss DL

EMDB-13549: 
Human coronavirus OC43 spike glycoprotein ectodomain in complex with the 46C12 antibody Fab fragment
Method: single particle / : Hesketh EL, Townend S, Ranson NA, Hurdiss DL

EMDB-13550: 
Human coronavirus OC43 spike glycoprotein ectodomain in complex with the 43E6 antibody Fab fragment
Method: single particle / : Hesketh EL, Townend S, Ranson NA, Hurdiss DL

EMDB-13563: 
Human coronavirus OC43 spike glycoprotein ectodomain in complex with the 47C9 antibody Fab fragment
Method: single particle / : Hesketh EL, Townend S, Ranson NA, Hurdiss DL

EMDB-13564: 
Human coronavirus OC43 spike glycoprotein ectodomain in complex with the 37F1 antibody Fab fragment
Method: single particle / : Hesketh EL, Townend S, Ranson NA, Hurdiss DL

PDB-7pnm: 
Human coronavirus OC43 spike glycoprotein ectodomain in complex with the 46C12 antibody Fab fragment
Method: single particle / : Hurdiss DL

PDB-7pnq: 
Human coronavirus OC43 spike glycoprotein ectodomain in complex with the 43E6 antibody Fab fragment
Method: single particle / : Hurdiss DL

PDB-7po5: 
Human coronavirus OC43 spike glycoprotein ectodomain in complex with the 47C9 antibody Fab fragment
Method: single particle / : Hurdiss DL

EMDB-12798: 
Hexameric coxsackievirus B3 2C protein in complex with S-fluoxetine
Method: single particle / : Hurdiss DL, Forster F

EMDB-21705: 
PL-bound rat TRPV2 in nanodiscs
Method: single particle / : Pumroy RP, Moiseenkova-Bell VY

PDB-6wkn: 
PL-bound rat TRPV2 in nanodiscs
Method: single particle / : Pumroy RP, Moiseenkova-Bell VY

EMDB-11514: 
Molecular pore spanning the double membranes of murine coronaviral (MHV-A59) replication organelles.
Method: subtomogram averaging / : Barcena M, Wolff G

EMDB-11515: 
Molecular pore spanning the double membranes of a mutant murine coronaviral (MHV-A59 GFP-nsp3) replication organelles.
Method: subtomogram averaging / : Barcena M, Wolff G
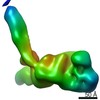
EMDB-10870: 
Cohesin complex with loader gripping DNA
Method: single particle / : Higashi TL, Eickhoff P, Sousa JS, Costa A, Uhlmann F

EMDB-10930: 
Cohesin complex with loader gripping DNA
Method: single particle / : Higashi TL, Eickhoff P, Sousa JS, Costa A, Uhlmann F

PDB-6yuf: 
Cohesin complex with loader gripping DNA
Method: single particle / : Higashi TL, Eickhoff P, Sousa JS, Costa A, Uhlmann F

EMDB-10676: 
Human Coronavirus HKU1 Haemagglutinin-Esterase
Method: single particle / : Hurdiss DL, Drulyte I

PDB-6y3y: 
Human Coronavirus HKU1 Haemagglutinin-Esterase
Method: single particle / : Hurdiss DL, Drulyte I, Pronker MF

EMDB-10337: 
XPF-ERCC1 Cryo-EM Structure, Apo-form
Method: single particle / : Jones ML, Briggs DC, McDonald NQ

EMDB-10338: 
XPF-ERCC1 Cryo-EM Structure, DNA-Bound form
Method: single particle / : Jones ML, Briggs DC, McDonald NQ

PDB-6sxa: 
XPF-ERCC1 Cryo-EM Structure, Apo-form
Method: single particle / : Jones ML, Briggs DC, McDonald NQ

PDB-6sxb: 
XPF-ERCC1 Cryo-EM Structure, DNA-Bound form
Method: single particle / : Jones ML, Briggs DC, McDonald NQ

EMDB-20584: 
A potent cross-neutralizing antibody targeting the fusion glycoprotein inhibits Nipah virus and Hendra virus infection
Method: single particle / : Dang HV, Chan YP, Park YJ, Snijder J, Da Silva SC, Vu B, Yan L, Feng YR, Rockx B, Geisbert T, Mire CE, Broder CB, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-6tys: 
A potent cross-neutralizing antibody targeting the fusion glycoprotein inhibits Nipah virus and Hendra virus infection
Method: single particle / : Dang HV, Chan YP, Park YJ, Snijder J, Da Silva SC, Vu B, Yan L, Feng YR, Rockx B, Geisbert T, Mire CE, Broder CB, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-0350: 
Induction of Potent Neutralizing Antibody Responses by a Designed Protein Nanoparticle Vaccine for Respiratory Syncytial Virus
Method: single particle / : Marcandalli J, Fiala B, Ols S, Perotti M, van der Schueren W, Snijder S, Hodge E, Benhaim M, Ravichandran R, Carter L, Sheffler W, Brunner L, Lawrenz M, Dubois P, Lanzavecchia A, Sallusto F, Lee KK, Veesler D, Correnti CE, Stewart LJ, Baker D, Lore K, Perez L, King NP

EMDB-0401: 
MERS-CoV complex with human neutralizing LCA60 antibody Fab fragment (state 1)
Method: single particle / : Walls AC, Xiong X, Park YJ, Tortorici MA, Snijder S, Quispe J, Cameroni E, Gopal R, Mian D, Lanzavecchia A, Zambon M, Rey FA, Corti D, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-0402: 
MERS-CoV S complex with human neutralizing LCA60 antibody Fab fragment (state 2)
Method: single particle / : Walls AC, Xiong X, Park YJ, Tortorici MA, Snijder S, Quispe J, Cameroni E, Gopal R, Mian D, Lanzavecchia A, Zambon M, Rey FA, Corti D, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-0403: 
SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 1)
Method: single particle / : Walls AC, Xiong X, Park YJ, Tortorici MA, Snijder S, Quispe J, Cameroni E, Gopal R, Mian D, Lanzavecchia A, Zambon M, Rey FA, Corti D, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-0404: 
SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 2)
Method: single particle / : Walls AC, Xiong X, Park YJ, Tortorici MA, Snijder S, Quispe J, Cameroni E, Gopal R, Mian D, Lanzavecchia A, Zambon M, Rey FA, Corti D, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-6nb3: 
MERS-CoV complex with human neutralizing LCA60 antibody Fab fragment (state 1)
Method: single particle / : Walls AC, Xiong X, Park YJ, Tortorici MA, Snijder S, Quispe J, Cameroni E, Gopal R, Mian D, Lanzavecchia A, Zambon M, Rey FA, Corti D, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-6nb4: 
MERS-CoV S complex with human neutralizing LCA60 antibody Fab fragment (state 2)
Method: single particle / : Walls AC, Xiong X, Park YJ, Tortorici MA, Snijder S, Quispe J, Cameroni E, Gopal R, Mian D, Lanzavecchia A, Zambon M, Rey FA, Corti D, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-6nb6: 
SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 1)
Method: single particle / : Walls AC, Xiong X, Park YJ, Tortorici MA, Snijder S, Quispe J, Cameroni E, Gopal R, Mian D, Lanzavecchia A, Zambon M, Rey FA, Corti D, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-6nb7: 
SARS-CoV complex with human neutralizing S230 antibody Fab fragment (state 2)
Method: single particle / : Walls AC, Xiong X, Park YJ, Tortorici MA, Snijder S, Quispe J, Cameroni E, Gopal R, Mian D, Lanzavecchia A, Zambon M, Rey FA, Corti D, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-9294: 
Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer, 3 Fabs bound, sharpened map
Method: single particle / : Borst AJ, Weidle CE, Gray MD, Frenz B, Snijder J, Joyce MG, Georgiev IS, Stewart-Jones GBE, Kwong PD, McGuire AT, DiMaio F, Stamatatos L, Pancera M, Veesler D
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
