-Search query
-Search result
Showing 1 - 50 of 165 items for (author: kim & hm)
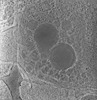
EMDB-28138: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY
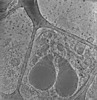
EMDB-28141: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY
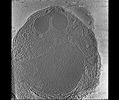
EMDB-28142: 
3D Reconstruction of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY

EMDB-34998: 
Cryo-EM structure of human TMEM87A, PE-bound
Method: single particle / : Han A, Kim HM

EMDB-35017: 
Cryo-EM structure of human TMEM87A, gluconate-bound
Method: single particle / : Han A, Kim HM

EMDB-18941: 
SARS-CoV-2 S (Spike) protein (BA.1) in complex with VHH Ma16B06 (sub-volume of two adjacent RBD-VHH modules)
Method: single particle / : Guttler T, Aksu M, Gorlich D

EMDB-40711: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-8sqn: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID)

EMDB-28728: 
Structure of 3A10 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase
Method: single particle / : Mou Z, Lei R, Wu NC, Dai X

EMDB-28729: 
Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase
Method: single particle / : Mou Z, Lei R, Wu NC, Dai X

EMDB-28730: 
Structure of 3C08 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase
Method: single particle / : Mou Z, Lei R, Wu NC, Dai X

EMDB-28125: 
Plasmodium falciparum merozoites apical 2-ring units
Method: subtomogram averaging / : Sun SY, Pintilie GD

EMDB-28126: 
Toxoplasma apical rings
Method: subtomogram averaging / : Sun SY, Pintilie GD

EMDB-41144: 
Cryo-EM Structure of GPR61-G protein complex stabilized by scFv16
Method: single particle / : Lees JA, Dias JM, Han S

EMDB-41145: 
Cryo-EM Structure of GPR61-
Method: single particle / : Lees JA, Dias JM, Han S

PDB-8tb0: 
Cryo-EM Structure of GPR61-G protein complex stabilized by scFv16
Method: single particle / : Lees JA, Dias JM, Han S

EMDB-29530: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-29531: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-40240: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-8fxb: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-8fxc: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-8s9g: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-40253: 
Truncated Braf/Mek/14-3-3 complex
Method: single particle / : Eck MJ, Jeon H, Park E, Rawson S

EMDB-34813: 
Structure of transforming growth factor beta induced protein (TGFBIp) G623R fibril
Method: helical / : Low JYK, Pervushin K

PDB-8hia: 
Structure of transforming growth factor beta induced protein (TGFBIp) G623R fibril
Method: helical / : Low JYK, Pervushin K

EMDB-27428: 
Cryo-EM structure of a RAS/RAF complex (state 1)
Method: single particle / : Eck MJ, Jeon H, Park E, Rawson S

EMDB-27429: 
Cryo-EM structure of a RAS/RAF complex (state 2)
Method: single particle / : Eck MJ, Jeon H, Park E, Rawson S

PDB-8dgs: 
Cryo-EM structure of a RAS/RAF complex (state 1)
Method: single particle / : Eck MJ, Jeon H, Park E, Rawson S

PDB-8dgt: 
Cryo-EM structure of a RAS/RAF complex (state 2)
Method: single particle / : Eck MJ, Jeon H, Park E, Rawson S

EMDB-35930: 
CryoEM structure of SARS CoV-2 RBD and Aptamer complex
Method: single particle / : Rahman MS, Jang SK, Lee JO

EMDB-35945: 
CryoEM structure of SARS CoV-2 RBD and Aptamer complex
Method: single particle / : Rahman MS, Jang SK, Lee JO

PDB-8j1q: 
CryoEM structure of SARS CoV-2 RBD and Aptamer complex
Method: single particle / : Rahman MS, Jang SK, Lee JO

PDB-8j26: 
CryoEM structure of SARS CoV-2 RBD and Aptamer complex
Method: single particle / : Rahman MS, Jang SK, Lee JO

EMDB-17540: 
Cryo-EM structure of full-length human UBR5 (homotetramer)
Method: single particle / : Aguirre JD, Kater L, Kempf G, Cavadini S, Thoma NH

PDB-8p83: 
Cryo-EM structure of full-length human UBR5 (homotetramer)
Method: single particle / : Aguirre JD, Kater L, Kempf G, Cavadini S, Thoma NH

EMDB-29533: 
Cryo-EM structure of Stanieria sp. CphA2
Method: single particle / : Markus LM, Sharon I, Strauss M, Schmeing TM

EMDB-29534: 
Cryo-EM structure of Stanieria sp. CphA2 in complex with ADPCP and 4x(beta-Asp-Arg)
Method: single particle / : Markus LM, Sharon I, Strauss M, Schmeing TM

PDB-8fxh: 
Cryo-EM structure of Stanieria sp. CphA2
Method: single particle / : Markus LM, Sharon I, Strauss M, Schmeing TM

PDB-8fxi: 
Cryo-EM structure of Stanieria sp. CphA2 in complex with ADPCP and 4x(beta-Asp-Arg)
Method: single particle / : Markus LM, Sharon I, Strauss M, Schmeing TM

EMDB-17542: 
Negative stain map of UBR5 (dimer) in complex with RARA/RXRA
Method: single particle / : Aguirre JD, Cavadini S, Kempf G, Kater L, Thoma NH

EMDB-28650: 
Human S1P transporter Spns2 in an inward-facing open conformation (state 1)
Method: single particle / : Ahmed S, Zhao H, Dai Y, Lee CH

EMDB-28651: 
Human S1P transporter Spns2 in an outward-facing open conformation (state 4)
Method: single particle / : Ahmed S, Zhao H, Dai Y, Lee CH

EMDB-28652: 
Human S1P transporter Spns2 in an inward-facing open conformation (state 1*)
Method: single particle / : Ahmed S, Zhao H, Dai Y, Lee CH

EMDB-28653: 
Human S1P transporter Spns2 in an outward-facing partially occluded conformation (state 3)
Method: single particle / : Ahmed S, Zhao H, Dai Y, Lee CH

EMDB-28654: 
Human S1P transporter Spns2 in an outward-facing partially occluded conformation (state 2)
Method: single particle / : Ahmed S, Zhao H, Dai Y, Lee CH

PDB-8ex4: 
Human S1P transporter Spns2 in an inward-facing open conformation (state 1)
Method: single particle / : Ahmed S, Zhao H, Dai Y, Lee CH

PDB-8ex5: 
Human S1P transporter Spns2 in an outward-facing open conformation (state 4)
Method: single particle / : Ahmed S, Zhao H, Dai Y, Lee CH

PDB-8ex6: 
Human S1P transporter Spns2 in an inward-facing open conformation (state 1*)
Method: single particle / : Ahmed S, Zhao H, Dai Y, Lee CH

PDB-8ex7: 
Human S1P transporter Spns2 in an outward-facing partially occluded conformation (state 3)
Method: single particle / : Ahmed S, Zhao H, Dai Y, Lee CH
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
