[English] 日本語
 Yorodumi
Yorodumi- EMDB-18941: SARS-CoV-2 S (Spike) protein (BA.1) in complex with VHH Ma16B06 (... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 S (Spike) protein (BA.1) in complex with VHH Ma16B06 (sub-volume of two adjacent RBD-VHH modules) | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  SARS-CoV-2 / SARS-CoV-2 /  Spike Glycoprotein / VHH Antibody (Nanobody) Complex / Inhibitor / Spike Glycoprotein / VHH Antibody (Nanobody) Complex / Inhibitor /  VIRAL PROTEIN VIRAL PROTEIN | |||||||||
| Biological species |   Severe acute respiratory syndrome coronavirus / Severe acute respiratory syndrome coronavirus /   Vicugna pacos (alpaca) Vicugna pacos (alpaca) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.6 Å cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Guttler T / Aksu M / Gorlich D | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Antiviral Res / Year: 2024 Journal: Antiviral Res / Year: 2024Title: Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters. Authors: Metin Aksu / Priya Kumar / Thomas Güttler / Waltraud Taxer / Kathrin Gregor / Bianka Mußil / Oleh Rymarenko / Kim M Stegmann / Antje Dickmanns / Sabrina Gerber / Wencke Reineking / Claudia ...Authors: Metin Aksu / Priya Kumar / Thomas Güttler / Waltraud Taxer / Kathrin Gregor / Bianka Mußil / Oleh Rymarenko / Kim M Stegmann / Antje Dickmanns / Sabrina Gerber / Wencke Reineking / Claudia Schulz / Timo Henneck / Ahmed Mohamed / Gerhard Pohlmann / Mehmet Ramazanoglu / Kemal Mese / Uwe Groß / Tamar Ben-Yedidia / Oded Ovadia / Dalit Weinstein Fischer / Merav Kamensky / Amir Reichman / Wolfgang Baumgärtner / Maren von Köckritz-Blickwede / Matthias Dobbelstein / Dirk Görlich /   Abstract: The ongoing threat of COVID-19 has highlighted the need for effective prophylaxis and convenient therapies, especially for outpatient settings. We have previously developed highly potent single- ...The ongoing threat of COVID-19 has highlighted the need for effective prophylaxis and convenient therapies, especially for outpatient settings. We have previously developed highly potent single-domain (VHH) antibodies, also known as nanobodies, that target the Receptor Binding Domain (RBD) of the SARS-CoV-2 Spike protein and neutralize the Wuhan strain of the virus. In this study, we present a new generation of anti-RBD nanobodies with superior properties. The primary representative of this group, Re32D03, neutralizes Alpha to Delta as well as Omicron BA.2.75; other members neutralize, in addition, Omicron BA.1, BA.2, BA.4/5, and XBB.1. Crystal structures of RBD-nanobody complexes reveal how ACE2-binding is blocked and also explain the nanobodies' tolerance to immune escape mutations. Through the cryo-EM structure of the Ma16B06-BA.1 Spike complex, we demonstrated how a single nanobody molecule can neutralize a trimeric spike. We also describe a method for large-scale production of these nanobodies in Pichia pastoris, and for formulating them into aerosols. Exposing hamsters to these aerosols, before or even 24 h after infection with SARS-CoV-2, significantly reduced virus load, weight loss and pathogenicity. These results show the potential of aerosolized nanobodies for prophylaxis and therapy of coronavirus infections. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18941.map.gz emd_18941.map.gz | 38.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18941-v30.xml emd-18941-v30.xml emd-18941.xml emd-18941.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18941_fsc.xml emd_18941_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_18941.png emd_18941.png | 107.9 KB | ||
| Masks |  emd_18941_msk_1.map emd_18941_msk_1.map | 42.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18941.cif.gz emd-18941.cif.gz | 7 KB | ||
| Others |  emd_18941_additional_1.map.gz emd_18941_additional_1.map.gz emd_18941_half_map_1.map.gz emd_18941_half_map_1.map.gz emd_18941_half_map_2.map.gz emd_18941_half_map_2.map.gz | 32.6 MB 32.8 MB 32.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18941 http://ftp.pdbj.org/pub/emdb/structures/EMD-18941 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18941 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18941 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18941.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18941.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18941_msk_1.map emd_18941_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
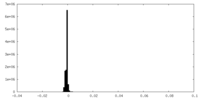
| Density Histograms |
-Additional map: unsharpened map
| File | emd_18941_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
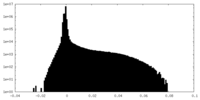
| Density Histograms |
-Half map: half map 2
| File | emd_18941_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_18941_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 S (Spike) protein (BA.1 variant) in complex with VHH M...
| Entire | Name: SARS-CoV-2 S (Spike) protein (BA.1 variant) in complex with VHH Ma16B06 (sub-volume of two adjacent RBD-VHH modules) |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 S (Spike) protein (BA.1 variant) in complex with VHH M...
| Supramolecule | Name: SARS-CoV-2 S (Spike) protein (BA.1 variant) in complex with VHH Ma16B06 (sub-volume of two adjacent RBD-VHH modules) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Trimeric SARS-CoV-2 S (Spike) protein (BA.1 variant) in complex with VHH antibody (nanobody) Ma16B06 (one copy per protomer). |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus / Strain: BA.1 Severe acute respiratory syndrome coronavirus / Strain: BA.1 |
| Molecular weight | Theoretical: 600 KDa |
-Macromolecule #1: SARS-CoV-2 HexaPro S (Spike) glycoprotein (BA.1)
| Macromolecule | Name: SARS-CoV-2 HexaPro S (Spike) glycoprotein (BA.1) / type: protein_or_peptide / ID: 1 Details: The Spike protein is a trimeric assembly. The map only shows two adjacent receptor-binding domains (RBDs), associated with one VHH antibody (Ma16B06) each. Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus / Strain: BA.1 Severe acute respiratory syndrome coronavirus / Strain: BA.1 |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VNLTTRTQLP PAYTNSFTRG VYYPDKVFRS SVLHSTQDLF LPFFSNVTWF HVISGTNGTK RFDNPVLPFN DGVYFASIEK SNIIRGWIFG TTLDSKTQSL LIVNNATNVV IKVCEFQFCN DPFLDHKNNK SWMESEFRVY SSANNCTFEY VSQPFLMDLE GKQGNFKNLR ...String: VNLTTRTQLP PAYTNSFTRG VYYPDKVFRS SVLHSTQDLF LPFFSNVTWF HVISGTNGTK RFDNPVLPFN DGVYFASIEK SNIIRGWIFG TTLDSKTQSL LIVNNATNVV IKVCEFQFCN DPFLDHKNNK SWMESEFRVY SSANNCTFEY VSQPFLMDLE GKQGNFKNLR EFVFKNIDGY FKIYSKHTPI IVREPEDLPQ GFSALEPLVD LPIGINITRF QTLLALHRSY LTPGDSSSGW TAGAAAYYVG YLQPRTFLLK YNENGTITDA VDCALDPLSE TKCTLKSFTV EKGIYQTSNF RVQPTESIVR FPNITNLCPF DEVFNATRFA SVYAWNRKRI SNCVADYSVL YNLAPFFTFK CYGVSPTKLN DLCFTNVYAD SFVIRGDEVR QIAPGQTGNI ADYNYKLPDD FTGCVIAWNS NKLDSKVSGN YNYLYRLFRK SNLKPFERDI STEIYQAGNK PCNGVAGFNC YFPLRSYSFR PTYGVGHQPY RVVVLSFELL HAPATVCGPK KSTNLVKNKC VNFNFNGLKG TGVLTESNKK FLPFQQFGRD IADTTDAVRD PQTLEILDIT PCSFGGVSVI TPGTNTSNQV AVLYQGVNCT EVPVAIHADQ LTPTWRVYST GSNVFQTRAG CLIGAEYVNN SYECDIPIGA GICASYQTQT KSPGSASSVA SQSIIAYTMS LGAENSVAYS NNSIAIPTNF TISVTTEILP VSMTKTSVDC TMYICGDSTE CSNLLLQYGS FCTQLKRALT GIAVEQDKNT QEVFAQVKQI YKTPPIKYFG GFNFSQILPD PSKPSKRSPI EDLLFNKVTL ADAGFIKQYG DCLGDIAARD LICAQKFKGL TVLPPLLTDE MIAQYTSALL AGTITSGWTF GAGPALQIPF PMQMAYRFNG IGVTQNVLYE NQKLIANQFN SAIGKIQDSL SSTPSALGKL QDVVNHNAQA LNTLVKQLSS KFGAISSVLN DIFSRLDPPE AEVQIDRLIT GRLQSLQTYV TQQLIRAAEI RASANLAATK MSECVLGQSK RVDFCGKGYH LMSFPQSAPH GVVFLHVTYV PAQEKNFTTA PAICHDGKAH FPREGVFVSN GTHWFVTQRN FYEPQIITTD NTFVSGNCDV VIGIVNNTVY DPLQPELDSF KEELDKYFKN HTSPDVDLGD ISGINASVVN IQKEIDRLNE VAKNLNESLI DLQELGKYEQ GSGYIPEAPR DGQAYVRKDG EWVLLSTFLG RSLEVLFQGP GHHHHHHHHS AWSHPQFEKG GGSGGGGSGG SAWSHPQFEK |
-Macromolecule #2: VHH antibody (nanobody) Ma16B06
| Macromolecule | Name: VHH antibody (nanobody) Ma16B06 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Vicugna pacos (alpaca) Vicugna pacos (alpaca) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: GSQVQLVESG GGLVRTGGSL RLSCAASGSI LQIWAMKWYR QAPGLQREWI ATIPNSGEPF YASSVEGRFT GSRENEETVY LYLNNLEPED TAVYYCEVNE GVPVREYWGQ GTQVTVSSTS |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | ||||||
| Details | monodisperse material eluted from a size exclusion column (Superose 6 Increase 3.2/300, Cytiva) |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Temperature | Min: 77.0 K / Max: 77.0 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 7592 / Average exposure time: 0.075 sec. / Average electron dose: 39.67 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller







 Z
Z Y
Y X
X