-Search query
-Search result
Showing 1 - 50 of 177 items for (author: johnson & je)

EMDB-28957: 
Cryo-EM structure of the Agrobacterium T-pilus
Method: helical / : Kreida S, Narita A, Johnson MD, Tocheva EI, Das A, Jensen GJ, Ghosal D

PDB-8fai: 
Cryo-EM structure of the Agrobacterium T-pilus
Method: helical / : Kreida S, Narita A, Johnson MD, Tocheva EI, Das A, Jensen GJ, Ghosal D

EMDB-42139: 
Cryo-EM structure of the flagellar MotAB stator bound to FliG
Method: single particle / : Deme JC, Johnson S, Lea SM

EMDB-42376: 
Cryo-EM structure of a single subunit of a Counterclockwise-locked form of the Salmonella enterica Typhimurium flagellar C-ring.
Method: single particle / : Johnson S, Deme JC, Lea SM

EMDB-42387: 
Cryo-EM structure of a single subunit of a Clockwise-locked form of the Salmonella enterica Typhimurium flagellar C-ring.
Method: single particle / : Johnson S, Deme JC, Lea SM

EMDB-42439: 
Cryo-EM structure of a Counterclockwise locked form of the Salmonella enterica Typhimurium flagellar C-ring, with C34 symmetry applied
Method: single particle / : Johnson S, Deme JC, Lea SM

EMDB-42451: 
Cryo-EM structure of a Clockwise locked form of the Salmonella enterica Typhimurium flagellar C-ring, with C34 symmetry applied
Method: single particle / : Johnson S, Deme JC, Lea SM

EMDB-29530: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-29531: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-40240: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-8fxb: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-8fxc: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-8s9g: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-41153: 
Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-41154: 
Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

PDB-8tcf: 
Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

PDB-8tcg: 
Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-29930: 
T. cruzi topoisomerase II alpha bound to dsDNA and the covalent inhibitor CT1
Method: single particle / : Schenk A, Deniston C, Noeske J

PDB-8gcc: 
T. cruzi topoisomerase II alpha bound to dsDNA and the covalent inhibitor CT1
Method: single particle / : Schenk A, Deniston C, Noeske J

EMDB-15084: 
cryo-EM structure of thioredoxin glutathione reductase in complex with a non-competitive inhibitor
Method: single particle / : Ardini M, Angelucci F, Fata F, Gabriele F, Effantin G, Ling W, Williams DL, Petukhova VZ, Petukhov PA

PDB-8a1r: 
cryo-EM structure of thioredoxin glutathione reductase in complex with a non-competitive inhibitor
Method: single particle / : Ardini M, Angelucci F, Fata F, Gabriele F, Effantin G, Ling W, Williams DL, Petukhova VZ, Petukhov PA

EMDB-15112: 
Nudaurelia capensis omega virus maturation intermediate captured at pH5.9 (insect cell expressed VLPs)
Method: single particle / : Castells-Graells R, Hesketh EL, Johnson JE, Ranson NA, Lawson DM, Lomonossoff GP

EMDB-15134: 
Nudaurelia capensis omega virus procapsid at pH7.6 (insect cell expressed VLPs)
Method: single particle / : Castells-Graells R, Hesketh EL, Johnson JE, Ranson NA, Lawson DM, Lomonossoff GP

EMDB-15209: 
Nudaurelia capensis omega virus maturation intermediate captured at pH6.25 (insect cell expressed VLPs)
Method: single particle / : Castells-Graells R, Hesketh EL, Johnson JE, Ranson NA, Lawson DM, Lomonossoff GP

EMDB-15266: 
Nudaurelia capensis omega virus maturation intermediates captured at pH5.6 (insect cell expressed VLPs): consensus map
Method: single particle / : Castells-Graells R, Hesketh EL, Johnson JE, Ranson NA, Lawson DM, Lomonossoff GP

EMDB-15307: 
Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): small class from symmetry expansion
Method: single particle / : Castells-Graells R, Hesketh EL, Johnson JE, Ranson NA, Lawson DM, Lomonossoff GP

EMDB-15339: 
Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): medium class from symmetry expansion
Method: single particle / : Castells-Graells R, Hesketh EL, Johnson JE, Ranson NA, Lawson DM, Lomonossoff GP

EMDB-15348: 
Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): large class from symmetry expansion
Method: single particle / : Castells-Graells R, Hesketh EL, Johnson JE, Ranson NA, Lawson DM, Lomonossoff GP
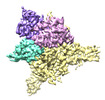
EMDB-27898: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA
Method: single particle / : Johnson ZL, Wasilko DJ, Ammirati M, Chang JS, Han S, Wu H
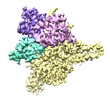
EMDB-27899: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA
Method: single particle / : Wasilko DJ, Johnson ZL, Ammirati M, Chang JS, Han S, Wu H

PDB-8e4y: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA
Method: single particle / : Johnson ZL, Wasilko DJ, Ammirati M, Chang JS, Han S, Wu H

PDB-8e50: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA
Method: single particle / : Wasilko DJ, Johnson ZL, Ammirati M, Chang JS, Han S, Wu H

EMDB-28596: 
CryoEM Structure of NLRP3 NACHT domain in complex with G2394
Method: single particle / : Murray JM, Johnson MC

PDB-8etr: 
CryoEM Structure of NLRP3 NACHT domain in complex with G2394
Method: single particle / : Murray JM, Johnson MC

EMDB-26507: 
SARS-CoV-2 spike in complex with Multivalent miniprotein inhibitor FUS231-P24 (2RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26508: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS231-P24 (3RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26509: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (2RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26510: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (3RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26511: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175 (local refinement of the RBD and AHB2)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26512: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-7uhb: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175 (local refinement of the RBD and AHB2)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-7uhc: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-25574: 
SARS-CoV-2 S-RBD + Fab 54042-4
Method: single particle / : Johnson NV, Mclellan JS

EMDB-26064: 
SARS-CoV-2 S + Fabs 5317-4 and 5217-10
Method: single particle / : Johnson NV, McLellan JS

EMDB-24437: 
HUMAN IMPDH1 TREATED WITH ATP, IMP, AND NAD+
Method: single particle / : Burrell AL, Kollman JM

EMDB-24438: 
HUMAN IMPDH1 TREATED WITH ATP, IMP, AND NAD+, OCTAMER-CENTERED
Method: single particle / : Burrell AL, Kollman JM

EMDB-24439: 
HUMAN IMPDH1 TREATED WITH GTP, IMP, AND NAD+; INTERFACE-CENTERED
Method: single particle / : Burrell AL, Kollman JM

EMDB-24440: 
HUMAN RETINAL VARIANT IMPDH1(595) TREATED WITH ATP; INTERFACE-CENTERED
Method: single particle / : Burrell AL, Kollman JM

EMDB-24441: 
HUMAN IMPDH1 TREATED WITH GTP, IMP, AND NAD+ OCTAMER-CENTERED
Method: single particle / : Burrell AL, Kollman JM

EMDB-24442: 
HUMAN RETINAL VARIANT IMPDH1(595) TREATED WITH ATP, OCTAMER-CENTERED
Method: single particle / : Burrell AL, Kollman JM
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
