-Search query
-Search result
Showing all 30 items for (author: erramilli & sk)
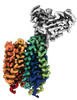
EMDB-43683: 
Cryo-EM structure of FLVCR2 in the inward-facing state with choline bound
Method: single particle / : Cater RJ, Mancia F
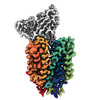
EMDB-43684: 
Cryo-EM structure of FLVCR2 in the outward-facing state with choline bound
Method: single particle / : Cater RJ, Mancia F
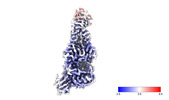
EMDB-40622: 
Chlorella virus Hyaluronan Synthase bound to GlcA extended GlcNAc primer
Method: single particle / : Stephens Z, Zimmer J

EMDB-40623: 
Chlorella virus Hyaluronan Synthase bound to GlcNAc primer and UDP-GlcA
Method: single particle / : Stephens Z, Zimmer J

EMDB-40624: 
Chlorella virus Hyaluronan Synthase bound to GlcA extended GlcNAc primer and UDP
Method: single particle / : Stephens Z, Zimmer J

EMDB-40591: 
Xenopus laevis hyaluronan synthase 1
Method: single particle / : Gorniak I, Zimmer J

EMDB-40594: 
Xenopus laevis hyaluronan synthase 1, nascent HA polymer bound state
Method: single particle / : Gorniak I, Zimmer J
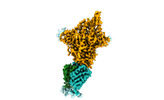
EMDB-40598: 
Xenopus laevis hyaluronan synthase 1, UDP-bound, gating loop inserted state
Method: single particle / : Gorniak I, Zimmer J
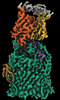
EMDB-41255: 
Cryo-EM structure of Pseudomonas aeruginosa TonB-dependent transporter PhuR in complex with synthetic antibody and heme
Method: single particle / : Knejski P, Erramilli SK, Kossiakoff AA

PDB-8the: 
Cryo-EM structure of Pseudomonas aeruginosa TonB-dependent transporter PhuR in complex with synthetic antibody and heme
Method: single particle / : Knejski P, Erramilli SK, Kossiakoff AA
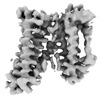
EMDB-24698: 
Cryo-EM structure of the HIV-1 restriction factor human SERINC3
Method: single particle / : Purdy MD, Leonhardt SA, Yeager M

EMDB-24705: 
Human SERINC3-DeltaICL4
Method: single particle / : Purdy MD, Leonhardt SA, Yeager M

EMDB-25836: 
CryoEM Structure of sFab COP-3 Complex with human claudin-4 and Clostridium perfringens enterotoxin C-terminal domain focused map
Method: single particle / : Vecchio AJ, Orlando BJ

EMDB-25750: 
Structure of the peroxisomal retro-translocon formed by a heterotrimeric ubiquitin ligase complex
Method: single particle / : Peiqiang F, Tom R

PDB-7t92: 
Structure of the peroxisomal retro-translocon formed by a heterotrimeric ubiquitin ligase complex
Method: single particle / : Peiqiang F, Tom R
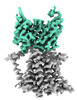
EMDB-26054: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state
Method: single particle / : Ashraf KU, Nygaard R, Vickery ON, Erramilli SK, Herrera CM, McConville TH, Petrou VI, Giacometti SI, Dufrisne MB, Nosol K, Zinkle AP, Graham CLB, Loukeris M, Kloss B, Skorupinska-Tudek K, Swiezewska E, Roper D, Clarke OB, Uhlemann AC, Kossiakoff AA, Trent MS, Stansfeld PJ, Mancia F
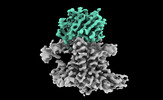
EMDB-26057: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state
Method: single particle / : Ashraf KU, Nygaard R, Vickery ON, Erramilli SK, Herrera CM, McConville TH, Petrou VI, Giacometti SI, Dufrisne MB, Nosol K, Zinkle AP, Graham CLB, Loukeris M, Kloss B, Skorupinska-Tudek K, Swiezewska E, Roper D, Clarke OB, Uhlemann AC, Kossiakoff AA, Trent MS, Stansfeld PJ, Mancia F

PDB-7tpg: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state
Method: single particle / : Ashraf KU, Nygaard R, Vickery ON, Erramilli SK, Herrera CM, McConville TH, Petrou VI, Giacometti SI, Dufrisne MB, Nosol K, Zinkle AP, Graham CLB, Loukeris M, Kloss B, Skorupinska-Tudek K, Swiezewska E, Roper D, Clarke OB, Uhlemann AC, Kossiakoff AA, Trent MS, Stansfeld PJ, Mancia F

PDB-7tpj: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state
Method: single particle / : Ashraf KU, Nygaard R, Vickery ON, Erramilli SK, Herrera CM, McConville TH, Petrou VI, Giacometti SI, Dufrisne MB, Nosol K, Zinkle AP, Graham CLB, Loukeris M, Kloss B, Skorupinska-Tudek K, Swiezewska E, Roper D, Clarke OB, Uhlemann AC, Kossiakoff AA, Trent MS, Stansfeld PJ, Mancia F

EMDB-25834: 
CryoEM Structure of sFab COP-2 Complex with human claudin-4 and Clostridium perfringens enterotoxin C-terminal domain
Method: single particle / : Vecchio AJ, Orlando BJ

EMDB-25835: 
CryoEM Structure of sFab COP-3 Complex with human claudin-4 and Clostridium perfringens enterotoxin C-terminal domain
Method: single particle / : Vecchio AJ, Orlando BJ

EMDB-12365: 
Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab and QA2-Fab (apo-inward-open conformation)
Method: single particle / : Nosol K, Locher KP

EMDB-12366: 
Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab and QA2-Fab (phosphatidylcholine-bound, occluded conformation)
Method: single particle / : Nosol K, Locher KP

EMDB-12367: 
Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab (posaconazole-bound, inward-open conformation)
Method: single particle / : Nosol K, Locher KP

EMDB-23883: 
Single-Particle Cryo-EM Structure of Major Facilitator Superfamily Domain containing 2A in complex with LPC-18:3
Method: single particle / : Cater RJ, Chua GL, Erramilli SK, Keener JE, Choy BC, Tokarz P, Chin CF, Quek DQY, Kloss B, Pepe JG, Parisi G, Wong BH, Clarke OB, Marty MT, Kossiakoff AA, Khelashvili G, Silver DL, Mancia F

EMDB-20857: 
CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor in complex with varenicline
Method: single particle / : Alvarez FJD, Mukherjee S, Han S, Ammirati M, Kossiakoff AA

EMDB-20863: 
CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor with varenicline in complex with anti-BRIL synthetic antibody BAK5
Method: single particle / : Alvarez FJD, Mukherjee S, Han S, Ammirati M, Kossiakoff AA

PDB-6ur8: 
CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor in complex with varenicline
Method: single particle / : Alvarez FJD, Mukherjee S, Han S, Ammirati M, Kossiakoff AA

PDB-6usf: 
CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor with varenicline in complex with anti-BRIL synthetic antibody BAK5
Method: single particle / : Alvarez FJD, Mukherjee S, Han S, Ammirati M, Kossiakoff AA

EMDB-20806: 
Single-Particle Cryo-EM Structure of Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) 7G8 Isoform
Method: single particle / : Kim J, Tan YZ, Wicht KJ, Erramilli SK, Dhingra SK, Okombo J, Vendome J, Hagenah LM, Giacometti SI, Warren AL, Nosol K, Roepe PD, Potter CS, Carragher B, Kossiakoff AA, Quick M, Fidock DA, Mancia F
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
