-Search query
-Search result
Showing 1 - 50 of 1,329 items for (author: ban & v)

EMDB-43751: 
TRPM7 structure in complex with anticancer agent CCT128930 in closed state
Method: single particle / : Nadezhdin KD, Sobolevsky AI

PDB-8w2l: 
TRPM7 structure in complex with anticancer agent CCT128930 in closed state
Method: single particle / : Nadezhdin KD, Sobolevsky AI

EMDB-40650: 
Phosphoinositide phosphate 3 kinase gamma
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40651: 
Phosphoinositide phosphate 3 kinase gamma bound with ATP
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40652: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40653: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and Gbetagamma
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40654: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and two Gbetagamma subunits in State 1
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-40655: 
Phosphoinositide phosphate 3 kinase gamma bound with ADP and two Gbetagamma subunits in State 2
Method: single particle / : Chen CL, Tesmer JJG, Bandekar SJ, Cash J

EMDB-41907: 
Computationally Designed, Expandable O4 Octahedral Handshake Nanocage
Method: single particle / : Weidle C, Borst A
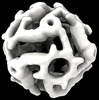
EMDB-42031: 
Computational Designed Nanocage O43_129_+8
Method: single particle / : Weidle C, Kibler RD

EMDB-19440: 
Cryo-EM structure of human NTCP-Bulevirtide complex
Method: single particle / : Liu H, Zakrzewicz D, Nosol K, Irobalieva RN, Mukherjee S, Bang-Soerensen R, Goldmann N, Kunz S, Rossi L, Kossiakoff AA, Urban S, Glebe D, Geyer J, Locher KP

PDB-8rqf: 
Cryo-EM structure of human NTCP-Bulevirtide complex
Method: single particle / : Liu H, Zakrzewicz D, Nosol K, Irobalieva RN, Mukherjee S, Bang-Soerensen R, Goldmann N, Kunz S, Rossi L, Kossiakoff AA, Urban S, Glebe D, Geyer J, Locher KP

EMDB-43318: 
Twistless helix 12 repeat ring design R12B
Method: single particle / : Calise SJ, Kollman JM

EMDB-29974: 
Cryo-EM structure of synthetic tetrameric building block sC4
Method: single particle / : Redler RL, Huddy TF, Hsia Y, Baker D, Ekiert D, Bhabha G

EMDB-41364: 
CryoEM Structure of a Computationally Designed T3 Tetrahedral Nanocage
Method: single particle / : Weidle C, Borst AJ

EMDB-42906: 
Computational Designed Nanocage O43_129
Method: single particle / : Weidle C, Kibler RD

EMDB-42944: 
Computational Designed Nanocage O43_129_+4
Method: single particle / : Carr KD, Weidle C, Borst AJ

PDB-8gel: 
Cryo-EM structure of synthetic tetrameric building block sC4
Method: single particle / : Redler RL, Huddy TF, Hsia Y, Baker D, Ekiert D, Bhabha G

PDB-8tl7: 
CryoEM Structure of a Computationally Designed T3 Tetrahedral Nanocage
Method: single particle / : Weidle C, Borst AJ

PDB-8v3b: 
Computational Designed Nanocage O43_129_+4
Method: single particle / : Carr KD, Weidle C, Borst AJ
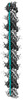
EMDB-19042: 
MAP7 MTBD (microtubule binding domain) decorated microtubule protofilament
Method: single particle / : Bangera M, Moores CA

EMDB-19043: 
MAP7 MTBD (microtubule binding domain) decorated Taxol stabilized microtubule (13pf) protofilament
Method: single particle / : Bangera M, Moores CA

EMDB-19044: 
MAP7 MTBD (microtubule binding domain) decorated Taxol stabilized microtubule (14pf) protofilament
Method: single particle / : Bangera M, Moores CA

PDB-8rc1: 
MAP7 MTBD (microtubule binding domain) decorated microtubule protofilament
Method: single particle / : Bangera M, Moores CA
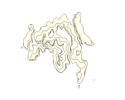
EMDB-19184: 
Late alpha-Synuclein fibril structure from liquid-liquid phase separations.
Method: helical / : De Simone A, Barritt JD, Chen S, Cascella R, Cecchi C, Bigi A, Jarvis JA, Chiti F, Dobson CM, Fusco G

PDB-8ri9: 
Late alpha-Synuclein fibril structure from liquid-liquid phase separations.
Method: helical / : De Simone A, Barritt JD, Chen S, Cascella R, Cecchi C, Bigi A, Jarvis JA, Chiti F, Dobson CM, Fusco G

EMDB-35817: 
Structure of Niacin-GPR109A-G protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

EMDB-35822: 
Structure of MK6892-GPR109A-G-protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

EMDB-35831: 
Structure of GSK256073-GPR109A-G-protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

EMDB-36193: 
Structure of Acipimox-GPR109A-G protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

EMDB-36280: 
Structure of MMF-GPR109A-G protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

PDB-8iy9: 
Structure of Niacin-GPR109A-G protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

PDB-8iyh: 
Structure of MK6892-GPR109A-G-protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

PDB-8iyw: 
Structure of GSK256073-GPR109A-G-protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

PDB-8jer: 
Structure of Acipimox-GPR109A-G protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

PDB-8jhn: 
Structure of MMF-GPR109A-G protein complex
Method: single particle / : Yadav MK, Sarma P, Chami M, Banerjee R, Shukla AK

EMDB-19250: 
Pseudoatomic model of a second-order Sierpinski triangle formed by the citrate synthase from Synechococcus elongatus
Method: single particle / : Lo YK, Bohn S, Sendker FL, Schuller JM, Hochberg G

EMDB-19251: 
Structure of a first order Sierpinski triangle formed by the H369R mutant of the citrate synthase from Synechococcus elongatus
Method: single particle / : Lo YK, Bohn S, Sendker FL, Schuller JM, Hochberg G

PDB-8rjk: 
Pseudoatomic model of a second-order Sierpinski triangle formed by the citrate synthase from Synechococcus elongatus
Method: single particle / : Lo YK, Bohn S, Sendker FL, Schuller JM, Hochberg G

PDB-8rjl: 
Structure of a first order Sierpinski triangle formed by the H369R mutant of the citrate synthase from Synechococcus elongatus
Method: single particle / : Lo YK, Bohn S, Sendker FL, Schuller JM, Hochberg G

EMDB-16903: 
60S ribosomal subunit bound to the E3-UFM1 complex (native, UFM1 pulldown)
Method: single particle / : Penchev I, DaRosa PA, Becker T, Beckmann R, Kopito R

EMDB-16880: 
60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (native)
Method: single particle / : Penchev I, DaRosa PA, Becker T, Beckmann R, Kopito R

EMDB-16902: 
60S ribosomal subunit bound to the E3-UFM1 complex - state 2 (native)
Method: single particle / : Penchev I, DaRosa PA, Becker T, Beckmann R, Kopito R

EMDB-16905: 
60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (in-vitro reconstitution)
Method: single particle / : Penchev I, DaRosa PA, Peter JJ, Kulathu Y, Becker T, Beckmann R, Kopito R

EMDB-16908: 
60S ribosomal subunit bound to the E3-UFM1 complex - state 1 (native)
Method: single particle / : Penchev I, DaRosa PA, Becker T, Beckmann R, Kopito R

PDB-8ohd: 
60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (native)
Method: single particle / : Penchev I, DaRosa PA, Becker T, Beckmann R, Kopito R

PDB-8oj0: 
60S ribosomal subunit bound to the E3-UFM1 complex - state 2 (native)
Method: single particle / : Penchev I, DaRosa PA, Becker T, Beckmann R, Kopito R

PDB-8oj5: 
60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (in-vitro reconstitution)
Method: single particle / : Penchev I, DaRosa PA, Peter JJ, Kulathu Y, Becker T, Beckmann R, Kopito R

PDB-8oj8: 
60S ribosomal subunit bound to the E3-UFM1 complex - state 1 (native)
Method: single particle / : Penchev I, DaRosa PA, Becker T, Beckmann R, Kopito R
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
