+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8v2d | ||||||
|---|---|---|---|---|---|---|---|
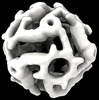
| Title | Computational Designed Nanocage O43_129 | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  DE NOVO PROTEIN / DE NOVO PROTEIN /  De Novo / De Novo /  Nanocage / O43_129 / extendable nanomaterials Nanocage / O43_129 / extendable nanomaterials | ||||||
| Biological species | synthetic construct (others) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 6.77 Å cryo EM / Resolution: 6.77 Å | ||||||
 Authors Authors | Weidle, C. / Kibler, R.D. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Blueprinting extendable nanomaterials with standardized protein blocks. Authors: Timothy F Huddy / Yang Hsia / Ryan D Kibler / Jinwei Xu / Neville Bethel / Deepesh Nagarajan / Rachel Redler / Philip J Y Leung / Connor Weidle / Alexis Courbet / Erin C Yang / Asim K Bera / ...Authors: Timothy F Huddy / Yang Hsia / Ryan D Kibler / Jinwei Xu / Neville Bethel / Deepesh Nagarajan / Rachel Redler / Philip J Y Leung / Connor Weidle / Alexis Courbet / Erin C Yang / Asim K Bera / Nicolas Coudray / S John Calise / Fatima A Davila-Hernandez / Hannah L Han / Kenneth D Carr / Zhe Li / Ryan McHugh / Gabriella Reggiano / Alex Kang / Banumathi Sankaran / Miles S Dickinson / Brian Coventry / T J Brunette / Yulai Liu / Justas Dauparas / Andrew J Borst / Damian Ekiert / Justin M Kollman / Gira Bhabha / David Baker /   Abstract: A wooden house frame consists of many different lumber pieces, but because of the regularity of these building blocks, the structure can be designed using straightforward geometrical principles. The ...A wooden house frame consists of many different lumber pieces, but because of the regularity of these building blocks, the structure can be designed using straightforward geometrical principles. The design of multicomponent protein assemblies, in comparison, has been much more complex, largely owing to the irregular shapes of protein structures. Here we describe extendable linear, curved and angled protein building blocks, as well as inter-block interactions, that conform to specified geometric standards; assemblies designed using these blocks inherit their extendability and regular interaction surfaces, enabling them to be expanded or contracted by varying the number of modules, and reinforced with secondary struts. Using X-ray crystallography and electron microscopy, we validate nanomaterial designs ranging from simple polygonal and circular oligomers that can be concentrically nested, up to large polyhedral nanocages and unbounded straight 'train track' assemblies with reconfigurable sizes and geometries that can be readily blueprinted. Because of the complexity of protein structures and sequence-structure relationships, it has not previously been possible to build up large protein assemblies by deliberate placement of protein backbones onto a blank three-dimensional canvas; the simplicity and geometric regularity of our design platform now enables construction of protein nanomaterials according to 'back of an envelope' architectural blueprints. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8v2d.cif.gz 8v2d.cif.gz | 1.8 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8v2d.ent.gz pdb8v2d.ent.gz | 1.2 MB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8v2d.json.gz 8v2d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/v2/8v2d https://data.pdbj.org/pub/pdb/validation_reports/v2/8v2d ftp://data.pdbj.org/pub/pdb/validation_reports/v2/8v2d ftp://data.pdbj.org/pub/pdb/validation_reports/v2/8v2d | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  42906MC  8g9jC  8g9kC  8ga6C  8ga7C  8gelC  8tl7C  8v3bC C: citing same article ( M: map data used to model this data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
|
 Movie
Movie Controller
Controller














 PDBj
PDBj