-Search query
-Search result
Showing 1 - 50 of 76 items for (author: baker & ml)

EMDB-18387: 
FtsH1 protease from P.aeruginosa clone C in negative stain
Method: single particle / : Mawla GD, Mansour Kamal S, Cao LY, Purhonen P, Hebert H, Sauer RT, Baker TA, Romling U

EMDB-18388: 
FtsH2 protease from P.aeruginosa clone C in negative stain
Method: single particle / : Mawla GD, Mansour Kamal S, Cao LY, Purhonen P, Hebert H, Sauer RT, Baker TA, Romling U

EMDB-18389: 
P.aeruginosa clone C construct PaFtsH2-H1-link32 in negative stain
Method: single particle / : Mawla GD, Mansour Kamal S, Cao LY, Purhonen P, Hebert H, Sauer RT, Baker TA, Romling U

EMDB-35245: 
Cryo-EM structure of the major capsid protein VP39 of Autographa californica multiple nucleopolyhedrovirus (AcMNPV)
Method: single particle / : Jia X, Zhang Q

EMDB-35246: 
Outer shell and inner layer structures of Autographa californica multiple nucleopolyhedrovirus (AcMNPV)
Method: single particle / : Jia X, Gao Y, Zhang Q

EMDB-35247: 
Plug structure of the Autographa californica multiple nucleopolyhedrovirus (AcMNPV)
Method: single particle / : Jia X, Gao Y, Zhang Q

EMDB-35242: 
The helical cylinder of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) nucleocapsid
Method: helical / : Jia X, Gao Y, Zhang Q

EMDB-35243: 
The apical part of Autographa californica multiple nucleopolyhedrovirus (AcMNPV)
Method: single particle / : Jia X, Gao Y, Zhang Q

EMDB-35244: 
The basal body of Autographa californica multiple nucleopolyhedrovirus (AcMNPV)
Method: single particle / : Jia X, Gao Y, Zhang Q

EMDB-41153: 
Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-41154: 
Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf
Method: single particle / : Campbell MG, Fernandez A, Roy A, Kraft J, Baker D

EMDB-27940: 
III2IV2 respiratory supercomplex from Saccharomyces cerevisiae with 4 bound UQ6
Method: single particle / : Hryc CF, Mileykovskaya E, Baker M, Dowhan W

EMDB-28011: 
III2IV respiratory supercomplex from Saccharomyces cerevisiae cardiolipin-lacking mutant
Method: single particle / : Hryc CF, Mileykovskaya E, Baker M, Dowhan W

EMDB-33403: 
3.1 Angstrom cryoEM icosahedral reconstruction of mud crab reovirus
Method: single particle / : Zhang Q, Gao Y

EMDB-33404: 
3.4 Angstrom cryoEM D5 reconstruction of mud crab reovirus
Method: single particle / : Zhang QF, Gao YZ

EMDB-33405: 
icosahedral reconstruction of mud crab reovirus in transcriptionally active state
Method: single particle / : Zhang Q, Gao Y

EMDB-33406: 
D5 reconstruction of mud crab reovirus in transcriptionally active state
Method: single particle / : Zhang Q, Gao Y
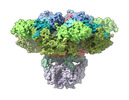
EMDB-27982: 
Structure of the full-length IP3R1 channel determined at high Ca2+
Method: single particle / : Fan G, Baker MR, Terry LE, Arige V, Chen M, Seryshev AB, Baker ML, Ludtke SJ, Yule DI, Serysheva II
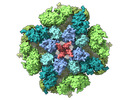
EMDB-27983: 
Structure of the full-length IP3R1 channel determined in the presence of Calcium/IP3/ATP
Method: single particle / : Fan G, Baker MR, Terry LE, Arige V, Chen M, Seryshev AB, Baker ML, Ludtke SJ, Yule DI, Serysheva II

EMDB-24609: 
Subnanometer in situ structure of the AcrAB-TolC efflux pump bound with MBX
Method: subtomogram averaging / : Muyuan MC, Zhao ZW

EMDB-24768: 
C1 reconstruction for Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by F-plasmid (pED208)
Method: single particle / : Liu X, Khara P, Baker ML, Christie PJ, Hu B

EMDB-24769: 
C13 reconstruction for Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by F-plasmid (pED208).
Method: single particle / : Liu X, Khara P, Baker ML, Christie PJ, Hu B

EMDB-24770: 
C17 reconstruction for Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by F-plasmid (pED208)
Method: single particle / : Liu X, Khara P, Baker ML, Christie PJ, Hu B

EMDB-24771: 
C13 reconstruction for Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by a plasmid overproducing TraV, TraK and TraB of pED208
Method: single particle / : Liu X, Khara P, Baker ML, Christie PJ, Hu B

EMDB-24772: 
C17 reconstruction for Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by a plasmid overproducing TraV, TraK and TraB of pED208
Method: single particle / : Liu X, Khara P, Baker ML, Christie PJ, Hu B

EMDB-24773: 
C16 reconstruction for Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by a plasmid overproducing TraV, TraK and TraB of pED208
Method: single particle / : Liu X, Khara P, Baker ML, Christie PJ, Hu B

EMDB-24408: 
The Capsid Structure of the ChAdOx1 viral vector/chimpanzee adenovirus Y25
Method: single particle / : Baker AT, Boyd RJ, Sarkar D, Vermaas JV, Williams D, Singharoy A

PDB-7rd1: 
The Capsid Structure of the ChAdOx1 viral vector/chimpanzee adenovirus Y25
Method: single particle / : Baker AT, Boyd RJ, Sarkar D, Vermaas JV, Williams D, Singharoy A

EMDB-23337: 
Structure of full-length IP3R1 channel reconstituted into lipid nanodisc in the apo-state
Method: single particle / : Baker MR, Fan G

EMDB-23338: 
Structure of full-length IP3R1 channel solubilized in LNMG & lipid in the apo-state
Method: single particle / : Baker MR, Fan G

EMDB-23265: 
Computationally designed icosahedral antibody nanocage with Fc i52.3+Fc
Method: single particle / : Dang HV, Veesler D

EMDB-23266: 
Computationally designed octahedral antibody nanocage with Fc o42.1+Fc
Method: single particle / : Dang HV, Veesler D

EMDB-9243: 
Structure of full-length IP3R1 channel bound with Adenophostin A (composite)
Method: single particle / : Serysheva II, Fan G, Baker MR, Wang Z, Seryshev A, Ludtke SJ, Baker ML

EMDB-9244: 
Structure of full-length IP3R1 channel in Apo-state (composite)
Method: single particle / : Serysheva II, Fan G, Baker MR, Wang Z, Seryshev A, Ludtke SJ, Baker ML

EMDB-9245: 
Structure of full-length IP3R1 channel in ligand-bound state
Method: single particle / : Fan G, Baker MR, Wang Z, Seryshev AB, Ludtke SJ, Baker ML, Serysheva II

EMDB-9246: 
Structure of full-length IP3R1 channel in Apo-state
Method: single particle / : Fan G, Baker MR, Wang Z, Seryshev AB, Ludtke SJ, Baker ML, Serysheva II

EMDB-9247: 
Structure of full-length IP3R1 channel in ligand-bound state
Method: single particle / : Fan G, Baker MR, Wang Z, Seryshev AB, Ludtke SJ, Baker ML, Serysheva II

EMDB-9248: 
Structure of full-length IP3R1 channel in Apo-state
Method: single particle / : Fan G, Baker MR, Wang Z, Seryshev AB, Ludtke SJ, Baker ML, Serysheva II

EMDB-6369: 
Structure of full-length IP3R1 channel in the apo-state determined by single particle cryo-EM
Method: single particle / : Fan G, Baker ML, Wang Z, Baker MR, Sinyagovskiy PA, Chiu W, Ludtke SJ, Serysheva II

EMDB-6258: 
Electron cryo-microscopy of the G protein effector, PDE6
Method: single particle / : Zhang Z, He F, Constantine R, Baker ML, Baehr W, Schmid MF, Wensel TG, Agosto MA

EMDB-5948: 
Three-dimensional structure of a protozoal dsRNA virus that infects enteric pathogen Giardia lamblia
Method: single particle / : Janssen ME, Takagi Y, Parent KN, Cardone G, Fichorova RN, Nibert ML, Baker TS

EMDB-6000: 
Full virus map of Brome Mosaic Virus
Method: single particle / : Wang Z, Hryc FC, Bammes B, Afonine PV, Jakana J, Chen DH, Liu XA, Baker ML, Kao C, Ludtke SJ, Schmid MF, Adams PD, Chiu W

EMDB-5954: 
Protruding Knob-like Proteins Violate Local Symmetries in an Icosahedral Marine Virus
Method: single particle / : Gipson P, Baker ML, Raytcheva D, Haase-Pettingell C, Piret J, King JA, Chiu W

EMDB-5725: 
Three-dimensional Structure of Victorivirus HvV190S
Method: single particle / : Dun SE, Li H, Cardone G, Nibert ML, Gabrial SA, Baker TS

EMDB-5726: 
Three-dimensional Structure of Victorivirus HvV190S
Method: single particle / : Dun SE, Li H, Cardone G, Nibert ML, Gabrial SA, Baker TS

EMDB-5727: 
Three-dimensional Structure of Victorivirus HvV190S
Method: single particle / : Dun SE, Li H, Cardone G, Nibert ML, Gabrial SA, Baker TS

EMDB-5678: 
Validated Near-Atomic Resolution Structure of Bacteriophage Epsilon15 Derived from Cryo-EM and Modeling
Method: single particle / : Baker ML, Hryc CF, Zhang Q, Wu W, Jakana J, Haase-Pettingell C, Afonine PV, Adams PD, King JA, Jiang W, Chiu W

EMDB-2184: 
Structure of protozoan virus from the obligate human genitourinary parasite Trichomonas vaginalis
Method: single particle / : Parent KN, Takagi Y, Cardone G, Olson NH, Ericsson M, Li Y, Fichorova RN, Nibert ML, Baker TS

EMDB-5275: 
4.4 Angstrom Cryo-EM Structure of an Enveloped Alphavirus Venezuelan Equine Encephalitis Virus
Method: single particle / : Zhang R, Hryc CF, Cong Y, Liu X, Jakana J, Gorchakov R, Baker ML, Weaver SC, Chiu W

EMDB-5276: 
4.4 Angstrom Cryo-EM Structure of an Enveloped Alphavirus Venezuelan Equine Encephalitis Virus
Method: single particle / : Zhang R, Hryc CF, Cong Y, Liu X, Jakana J, Gorchakov R, Baker ML, Weaver SC, Chiu W
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
