[English] 日本語
 Yorodumi
Yorodumi- EMDB-24408: The Capsid Structure of the ChAdOx1 viral vector/chimpanzee adeno... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24408 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The Capsid Structure of the ChAdOx1 viral vector/chimpanzee adenovirus Y25 | |||||||||
 Map data Map data | Relion-generated, post-processed map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationhexon binding /  viral capsid, decoration / T=25 icosahedral viral capsid / lysis of host organelle involved in viral entry into host cell / viral procapsid / microtubule-dependent intracellular transport of viral material towards nucleus / viral release from host cell / endocytosis involved in viral entry into host cell / viral capsid, decoration / T=25 icosahedral viral capsid / lysis of host organelle involved in viral entry into host cell / viral procapsid / microtubule-dependent intracellular transport of viral material towards nucleus / viral release from host cell / endocytosis involved in viral entry into host cell /  viral capsid / host cell cytoplasm ...hexon binding / viral capsid / host cell cytoplasm ...hexon binding /  viral capsid, decoration / T=25 icosahedral viral capsid / lysis of host organelle involved in viral entry into host cell / viral procapsid / microtubule-dependent intracellular transport of viral material towards nucleus / viral release from host cell / endocytosis involved in viral entry into host cell / viral capsid, decoration / T=25 icosahedral viral capsid / lysis of host organelle involved in viral entry into host cell / viral procapsid / microtubule-dependent intracellular transport of viral material towards nucleus / viral release from host cell / endocytosis involved in viral entry into host cell /  viral capsid / host cell cytoplasm / symbiont entry into host cell / host cell nucleus / virion attachment to host cell / structural molecule activity viral capsid / host cell cytoplasm / symbiont entry into host cell / host cell nucleus / virion attachment to host cell / structural molecule activitySimilarity search - Function | |||||||||
| Biological species |  Chimpanzee adenovirus Y25 Chimpanzee adenovirus Y25 | |||||||||
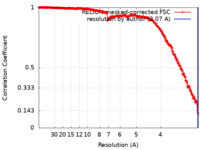
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.07 Å cryo EM / Resolution: 3.07 Å | |||||||||
 Authors Authors | Baker AT / Boyd RJ / Sarkar D / Vermaas JV / Williams D / Singharoy A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: ChAdOx1 interacts with CAR and PF4 with implications for thrombosis with thrombocytopenia syndrome. Authors: Alexander T Baker / Ryan J Boyd / Daipayan Sarkar / Alicia Teijeira-Crespo / Chun Kit Chan / Emily Bates / Kasim Waraich / John Vant / Eric Wilson / Chloe D Truong / Magdalena Lipka-Lloyd / ...Authors: Alexander T Baker / Ryan J Boyd / Daipayan Sarkar / Alicia Teijeira-Crespo / Chun Kit Chan / Emily Bates / Kasim Waraich / John Vant / Eric Wilson / Chloe D Truong / Magdalena Lipka-Lloyd / Petra Fromme / Josh Vermaas / Dewight Williams / LeeAnn Machiesky / Meike Heurich / Bolni M Nagalo / Lynda Coughlan / Scott Umlauf / Po-Lin Chiu / Pierre J Rizkallah / Taylor S Cohen / Alan L Parker / Abhishek Singharoy / Mitesh J Borad /   Abstract: Vaccines derived from chimpanzee adenovirus Y25 (ChAdOx1), human adenovirus type 26 (HAdV-D26), and human adenovirus type 5 (HAdV-C5) are critical in combatting the severe acute respiratory ...Vaccines derived from chimpanzee adenovirus Y25 (ChAdOx1), human adenovirus type 26 (HAdV-D26), and human adenovirus type 5 (HAdV-C5) are critical in combatting the severe acute respiratory coronavirus 2 (SARS-CoV-2) pandemic. As part of the largest vaccination campaign in history, ultrarare side effects not seen in phase 3 trials, including thrombosis with thrombocytopenia syndrome (TTS), a rare condition resembling heparin-induced thrombocytopenia (HIT), have been observed. This study demonstrates that all three adenoviruses deployed as vaccination vectors versus SARS-CoV-2 bind to platelet factor 4 (PF4), a protein implicated in the pathogenesis of HIT. We have determined the structure of the ChAdOx1 viral vector and used it in state-of-the-art computational simulations to demonstrate an electrostatic interaction mechanism with PF4, which was confirmed experimentally by surface plasmon resonance. These data confirm that PF4 is capable of forming stable complexes with clinically relevant adenoviruses, an important step in unraveling the mechanisms underlying TTS. #1:  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: The Structure of ChAdOx1/AZD-1222 Reveals Interactions with CAR and PF4 with Implications for Vaccine-induced Immune Thrombotic Thrombocytopenia Authors: Baker AT / Boyd RJ / Sarkar D / Vant J / Crespo AT / Waraich K / Truong CD / Bates E / Wilson E / Chan CK / Lipka-Lloyd M / Fromme P / Nagalo MB / Heurich M / Williams D / Chiu PL / ...Authors: Baker AT / Boyd RJ / Sarkar D / Vant J / Crespo AT / Waraich K / Truong CD / Bates E / Wilson E / Chan CK / Lipka-Lloyd M / Fromme P / Nagalo MB / Heurich M / Williams D / Chiu PL / Rizkallah PJ / Parker AL / Singharoy A / Borad MJ | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24408.map.gz emd_24408.map.gz | 715.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24408-v30.xml emd-24408-v30.xml emd-24408.xml emd-24408.xml | 36.5 KB 36.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24408_fsc.xml emd_24408_fsc.xml | 44 KB | Display |  FSC data file FSC data file |
| Images |  emd_24408.png emd_24408.png | 53.3 KB | ||
| Masks |  emd_24408_msk_1.map emd_24408_msk_1.map | 7.5 GB |  Mask map Mask map | |
| Others |  emd_24408_additional_1.map.gz emd_24408_additional_1.map.gz emd_24408_additional_2.map.gz emd_24408_additional_2.map.gz emd_24408_additional_3.map.gz emd_24408_additional_3.map.gz emd_24408_additional_4.map.gz emd_24408_additional_4.map.gz emd_24408_half_map_1.map.gz emd_24408_half_map_1.map.gz emd_24408_half_map_2.map.gz emd_24408_half_map_2.map.gz | 6.9 GB 6.2 GB 6.9 GB 312.1 MB 6.1 GB 6.1 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24408 http://ftp.pdbj.org/pub/emdb/structures/EMD-24408 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24408 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24408 | HTTPS FTP |
-Related structure data
| Related structure data |  7rd1MC  7op2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10754 (Title: Cryo TEM single particle dataset of purified ChAdOx1/ADZ-1222 EMPIAR-10754 (Title: Cryo TEM single particle dataset of purified ChAdOx1/ADZ-1222Data size: 1.0 TB Data #1: Raw movies comprising 7RD1 ChAdOx dataset [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24408.map.gz / Format: CCP4 / Size: 7.5 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24408.map.gz / Format: CCP4 / Size: 7.5 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion-generated, post-processed map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.53143 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_24408_msk_1.map emd_24408_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
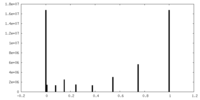
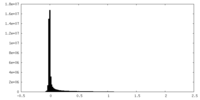
| Density Histograms |
-Additional map: DeepEMhancer-sharpened using highRes model
| File | emd_24408_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer-sharpened using highRes model | ||||||||||||
| Projections & Slices |
| ||||||||||||
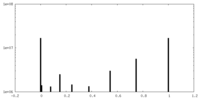
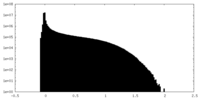
| Density Histograms |
-Additional map: DeepEMhancer-sharpened using wideTarget model
| File | emd_24408_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer-sharpened using wideTarget model | ||||||||||||
| Projections & Slices |
| ||||||||||||
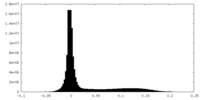
| Density Histograms |
-Additional map: DeepEMhancer-sharpened using wideTarget model and low pass-filtered
| File | emd_24408_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer-sharpened using wideTarget model and low pass-filtered | ||||||||||||
| Projections & Slices |
| ||||||||||||
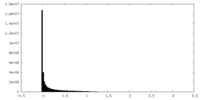
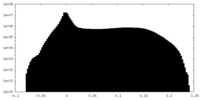
| Density Histograms |
-Additional map: Hollowed to remove noise generated by nonstructured genome
| File | emd_24408_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Hollowed to remove noise generated by nonstructured genome | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Random half map
| File | emd_24408_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Random half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
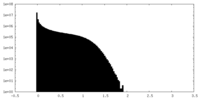
| Density Histograms |
-Half map: Random half map
| File | emd_24408_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Random half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Chimpanzee adenovirus Y25
| Entire | Name:  Chimpanzee adenovirus Y25 Chimpanzee adenovirus Y25 |
|---|---|
| Components |
|
-Supramolecule #1: Chimpanzee adenovirus Y25
| Supramolecule | Name: Chimpanzee adenovirus Y25 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Rescued from a bacterial artificial chromosome containing the ChAdOx1 genome and propagated in HEK293 T-Rex cells. NCBI-ID: 1123958 / Sci species name: Chimpanzee adenovirus Y25 / Sci species strain: ChAdOx1 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:   Pan troglodytes (chimpanzee) Pan troglodytes (chimpanzee) |
| Host system | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Virus shell | Shell ID: 1 / Name: Icosahedral Capsid / Diameter: 500.0 Å / T number (triangulation number): 25 |
-Macromolecule #1: Pre-protein VI
| Macromolecule | Name: Pre-protein VI / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chimpanzee adenovirus Y25 Chimpanzee adenovirus Y25 |
| Molecular weight | Theoretical: 26.299689 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEDINFSSLA PRHGTRPFMG TWSDIGTSQL NGGAFNWSSL WSGLKNFGST LKTYGSKAWN STTGQALRDK LKEQNFQQKV VDGLASGIN GVVDLANQAV QRQINSRLDP VPPAGSVEMP QVEEELPPLD KRGEKRPRPD AEETLLTHTD EPPPYEEAVK L GLPTTRPI ...String: MEDINFSSLA PRHGTRPFMG TWSDIGTSQL NGGAFNWSSL WSGLKNFGST LKTYGSKAWN STTGQALRDK LKEQNFQQKV VDGLASGIN GVVDLANQAV QRQINSRLDP VPPAGSVEMP QVEEELPPLD KRGEKRPRPD AEETLLTHTD EPPPYEEAVK L GLPTTRPI APLATGVLKP ESNKPATLDL PPPASRPSTV AKPLPPVAVA RARPGGSARP HANWQSTLNS IVGLGVQSVK RR RCY |
-Macromolecule #2: Hexon protein
| Macromolecule | Name: Hexon protein / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chimpanzee adenovirus Y25 Chimpanzee adenovirus Y25 |
| Molecular weight | Theoretical: 106.121141 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MATPSMLPQW AYMHIAGQDA SEYLSPGLVQ FARATDTYFS LGNKFRNPTV APTHDVTTDR SQRLTLRFVP VDREDNTYSY KVRYTLAVG DNRVLDMAST YFDIRGVLDR GPSFKPYSGT AYNSLAPKGA PNSSQWEQKK AGNGDTMETH TFGVAPMGGE N ITIDGLQI ...String: MATPSMLPQW AYMHIAGQDA SEYLSPGLVQ FARATDTYFS LGNKFRNPTV APTHDVTTDR SQRLTLRFVP VDREDNTYSY KVRYTLAVG DNRVLDMAST YFDIRGVLDR GPSFKPYSGT AYNSLAPKGA PNSSQWEQKK AGNGDTMETH TFGVAPMGGE N ITIDGLQI GTDATADQDK PIYADKTFQP EPQVGEENWQ ETESFYGGRA LKKDTSMKPC YGSYARPTNV KGGQAKLKVG AD GVPTKEF DIDLAFFDTP GGTVNGQDEY KADIVMYTEN TYLETPDTHV VYKPGKDDAS SEINLVQQSM PNRPNYIGFR DNF IGLMYY NSTGNMGVLA GQASQLNAVV DLQDRNTELS YQLLLDSLGD RTRYFSMWNQ AVDSYDPDVR IIENHGVEDE LPNY CFPLD GSGTNAAYQG VKVKNGNDGD VESEWENDDT VAARNQLCKG NIFAMEINLQ ANLWRSFLYS NVALYLPDSY KYTPA NITL PTNTNTYDYM NGRVVPPSLV DAYINIGARW SLDPMDNVNP FNHHRNAGLR YRSMLLGNGR YVPFHIQVPQ KFFAIK SLL LLPGSYTYEW NFRKDVNMIL QSSLGNDLRT DGASISFTSI NLYATFFPMA HNTASTLEAM LRNDTNDQSF NDYLSAA NM LYPIPANATN VPISIPSRNW AAFRGWSFTR LKTKETPSLG SGFDPYFVYS GSIPYLDGTF YLNHTFKKVS ITFDSSVS W PGNDRLLTPN EFEIKRTVDG EGYNVAQCNM TKDWFLVQML AHYNIGYQGF YVPEGYKDRM YSFFRNFQPM SRQVVDEVN YKDYQAVTLA YQHNNSGFVG YLAPTMRQGQ PYPANYPYPL IGKSAVTSVT QKKFLCDRVM WRIPFSSNFM SMGALTDLGQ NMLYANSAH ALDMNFEVDP MDESTLLYVV FEVFDVVRVH QPHRGVIEAV YLRTPFSAGN ATT |
-Macromolecule #3: Penton protein
| Macromolecule | Name: Penton protein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chimpanzee adenovirus Y25 Chimpanzee adenovirus Y25 |
| Molecular weight | Theoretical: 59.533656 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MMRRAYPEGP PPSYESVMQQ AMAAAAAMQP PLEAPYVPPR YLAPTEGRNS IRYSELAPLY DTTRLYLVDN KSADIASLNY QNDHSNFLT TVVQNNDFTP TEASTQTINF DERSRWGGQL KTIMHTNMPN VNEFMYSNKF KARVMVSRKT PNGVTVTDGS Q DILEYEWV ...String: MMRRAYPEGP PPSYESVMQQ AMAAAAAMQP PLEAPYVPPR YLAPTEGRNS IRYSELAPLY DTTRLYLVDN KSADIASLNY QNDHSNFLT TVVQNNDFTP TEASTQTINF DERSRWGGQL KTIMHTNMPN VNEFMYSNKF KARVMVSRKT PNGVTVTDGS Q DILEYEWV EFELPEGNFS VTMTIDLMNN AIIDNYLAVG RQNGVLESDI GVKFDTRNFR LGWDPVTELV MPGVYTNEAF HP DIVLLPG CGVDFTESRL SNLLGIRKRQ PFQEGFQIMY EDLEGGNIPA LLDVDAYEKS KEESAAAATA AVATASTEVR GDN FASPAA VAAAEAAETE SKIVIQPVEK DSKDRSYNVL PDKINTAYRS WYLAYNYGDP EKGVRSWTLL TTSDVTCGVE QVYW SLPDM MQDPVTFRST RQVSNYPVVG AELLPVYSKS FFNEQAVYSQ QLRAFTSLTH VFNRFPENQI LVRPPAPTIT TVSEN VPAL TDHGTLPLRS SIRGVQRVTV TDARRRTCPY VYKALGIVAP RVLSSRTF |
-Macromolecule #4: Hexon-interlacing protein
| Macromolecule | Name: Hexon-interlacing protein / type: protein_or_peptide / ID: 4 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chimpanzee adenovirus Y25 Chimpanzee adenovirus Y25 |
| Molecular weight | Theoretical: 14.468118 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSGSGSFEGG VFSPYLTGRL PSWAGVRQNV MGSTVDGRPV QPANSSTLTY ATLSSSSVDA AAAAAAASAA SAVRGMAMGA GYYGTLVAN SSSTNNPASL NEEKLLLLMA QLEALTQRLG ELTQQVAQLQ EQTRAAVATV KSK |
-Macromolecule #5: Pre-hexon-linking protein IIIa
| Macromolecule | Name: Pre-hexon-linking protein IIIa / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chimpanzee adenovirus Y25 Chimpanzee adenovirus Y25 |
| Molecular weight | Theoretical: 65.9545 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MQQQPPPDPA MRAALQSQPS GINSSDDWTQ AMQRIMALTT RNPEAFRQQP QANRLSAILE AVVPSRSNPT HEKVLAIVNA LVENKAIRG DEAGLVYNAL LERVARYNST NVQTNLDRMV TDVREAVAQR ERFHRESNLG SMVALNAFLS TQPANVPRGQ E DYTNFISA ...String: MQQQPPPDPA MRAALQSQPS GINSSDDWTQ AMQRIMALTT RNPEAFRQQP QANRLSAILE AVVPSRSNPT HEKVLAIVNA LVENKAIRG DEAGLVYNAL LERVARYNST NVQTNLDRMV TDVREAVAQR ERFHRESNLG SMVALNAFLS TQPANVPRGQ E DYTNFISA LRLMVTEVPQ SEVYQSGPDY FFQTSRQGLQ TVNLSQAFKN LQGLWGVQAP VGDRATVSSL LTPNSRLLLL LV APFTDSG SINRNSYLGY LINLYREAIG QAHVDEQTYQ EITHVSRALG QDDPGNLEAT LNFLLTNRSQ KIPPQYTLSA EEE RILRYV QQSVGLFLMQ EGATPSAALD MTARNMEPSM YASNRPFINK LMDYLHRAAA MNSDYFTNAI LNPHWLPPPG FYTG EYDMP DPNDGFLWDD VDSSVFSPRP GANERPLWKK EGSDRRPSSA LSGREGAAAA VPEAASPFPS LPFSLNSIRS SELGR ITRP RLLGEEEYLN DSLLRPEREK NFPNNGIESL VDKMSRWKTY AQEHRDDPSQ GATSRGSAAR KRRWHDRQRG LMWDDE DSA DDSSVLDLGG SGNPFAHLRP RIGRMM |
-Macromolecule #6: Pre-hexon-linking protein VIII
| Macromolecule | Name: Pre-hexon-linking protein VIII / type: protein_or_peptide / ID: 6 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chimpanzee adenovirus Y25 Chimpanzee adenovirus Y25 |
| Molecular weight | Theoretical: 24.779684 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSKEIPTPYM WSYQPQMGLA AGAAQDYSTR MNWLSAGPAM ISRVNDIRAH RNQILLEQSA LTATPRNHLN PRNWPAALVY QEIPQPTTV LLPRDAQAEV QLTNSGVQLA GGATLCRHRP AQGIKRLVIR GRGTQLNDEV VSSSLGLRPD GVFQLAGSGR S SFTPRQAV ...String: MSKEIPTPYM WSYQPQMGLA AGAAQDYSTR MNWLSAGPAM ISRVNDIRAH RNQILLEQSA LTATPRNHLN PRNWPAALVY QEIPQPTTV LLPRDAQAEV QLTNSGVQLA GGATLCRHRP AQGIKRLVIR GRGTQLNDEV VSSSLGLRPD GVFQLAGSGR S SFTPRQAV LTLESSSSQP RSGGIGTLQF VEEFTPSVYF NPFSGSPGHY PDEFIPNFDA ISESVDGYD |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil R2/2 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: Grid was R2/2 UltrAUFoil. Sample was applied and blotted twice before plunging.. | ||||||||||||
| Details | Concentrated by centrifugation |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 2875 / Average exposure time: 0.2 sec. / Average electron dose: 1.1802 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7rd1: |
 Movie
Movie Controller
Controller












 Z
Z Y
Y X
X