4RLB
 
 | |
4RL9
 
 | |
4RLC
 
 | |
6HDY
 
 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in the postadenylation state in complex with S3-HB-AMP | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, 2-hydroxyisobutyryl-CoA synthetase, SULFATE ION, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
6HDX
 
 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in the postadenylation state in complex with R3-HIB-AMP | | Descriptor: | (2R)-3-HYDROXY-2-METHYLPROPANOIC ACID, 2-hydroxyisobutyryl-CoA synthetase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] (2~{R})-2-methyl-3-oxidanyl-propanoate | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
6HE2
 
 | | Crystal structure of an open conformation of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in complex with 2-HIB-AMP and CoA | | Descriptor: | 2-hydroxyisobutyryl-CoA synthetase, ADENOSINE MONOPHOSPHATE, COENZYME A, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
6HE0
 
 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in complex with 2-HIB-AMP and CoA in the thioesterfication state | | Descriptor: | 2-hydroxyisobutyryl-CoA synthetase, ADENOSINE MONOPHOSPHATE, COENZYME A, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
5DL8
 
 | |
5DL5
 
 | |
5DL7
 
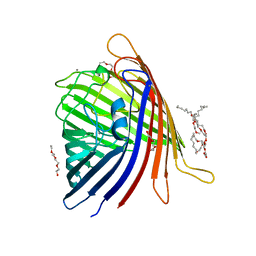 | |
5DL6
 
 | |
7PT4
 
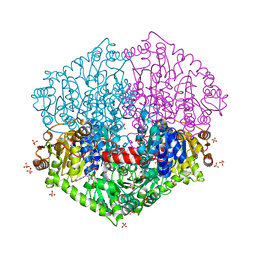 | | Actinobacterial 2-hydroxyacyl-CoA lyase (AcHACL) structure in complex with a covalently bound reaction intermediate as well as products formyl-CoA and acetone | | Descriptor: | 2-hydroxyacyl-CoA lyase, 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1R,11R,15S,17R)-19-[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]-1,11,15,17-TETRAHYDROXY-12,12-DIMETHYL-15,17-DIOXIDO-6,10-DIOXO-14,16,18-TRIOXA-2-THIA-5,9-DIAZA-15,17-DIPHOSPHANONADEC-1-YL}-5-(2-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, ACETONE, ... | | Authors: | Zahn, M, Rohwerder, T. | | Deposit date: | 2021-09-25 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanistic details of the actinobacterial lyase-catalyzed degradation reaction of 2-hydroxyisobutyryl-CoA.
J.Biol.Chem., 298, 2022
|
|
7PT1
 
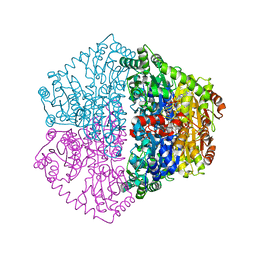 | |
7PT3
 
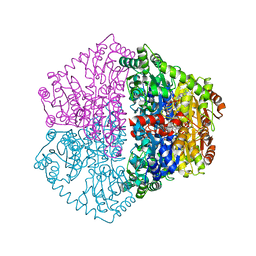 | |
7PT2
 
 | |
6HDW
 
 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in the postadenylation state in complex with 2-HIB-AMP | | Descriptor: | 2-hydroxyisobutyryl-CoA synthetase, SULFATE ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] 2-methyl-2-oxidanyl-propanoate | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
4EZP
 
 | |
4F01
 
 | |
4EZT
 
 | |
4EZR
 
 | |
4EZW
 
 | |
4EZO
 
 | |
4EZZ
 
 | |
4F00
 
 | |
4EZQ
 
 | |
