1N69
 
 | | Crystal structure of human saposin B | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, SAPOSIN B | | Authors: | Ahn, V.E, Faull, K.F, Whitelegge, J.P, Fluharty, A.L, Prive, G.G. | | Deposit date: | 2002-11-08 | | Release date: | 2003-01-07 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of saposin B reveals a dimeric shell for lipid binding
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4UNT
 
 | | Induced monomer of the Mcg variable domain | | Descriptor: | IG LAMBDA CHAIN V-II REGION MGC, SULFATE ION | | Authors: | Brumshtein, B, Esswein, S.R, Landau, M, Ryan, C.M, Whitelegge, J.P, Phillips, M.L, Cascio, D, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2014-05-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Formation of Amyloid Fibers by Monomeric Light-Chain Variable Domains.
J.Biol.Chem., 289, 2014
|
|
1Q5I
 
 | | Crystal structure of bacteriorhodopsin mutant P186A crystallized from bicelles | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Yohannan, S, Faham, S, Yang, D, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-08-07 | | Release date: | 2004-01-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The evolution of transmembrane helix kinks and the structural diversity of G protein-coupled receptors.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1PXR
 
 | | Structure of Pro50Ala mutant of Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Faham, S, Yang, D, Bare, E, Yohannan, S, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-07-06 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Side-chain Contributions to Membrane Protein Structure and Stability.
J.Mol.Biol., 335, 2004
|
|
1PY6
 
 | | Bacteriorhodopsin crystallized from bicells | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Faham, S, Yang, D, Bare, E, Yohannan, S, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-07-08 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Side-chain Contributions to Membrane Protein Structure and Stability.
J.Mol.Biol., 335, 2004
|
|
1PXS
 
 | | Structure of Met56Ala mutant of Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Faham, S, Yang, D, Bare, E, Yohannan, S, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-07-06 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Side-chain Contributions to Membrane Protein Structure and Stability.
J.Mol.Biol., 335, 2004
|
|
1Q5J
 
 | | Crystal structure of bacteriorhodopsin mutant P91A crystallized from bicelles | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Yohannan, S, Faham, S, Yang, D, Whitelegge, J.P, Bowie, J.U. | | Deposit date: | 2003-08-07 | | Release date: | 2004-01-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The evolution of transmembrane helix kinks and the structural diversity of G protein-coupled receptors.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8SZX
 
 | |
3MAY
 
 | |
2ZT9
 
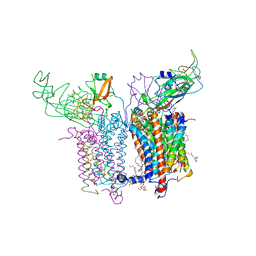 | | Crystal Structure of the Cytochrome b6f Complex from Nostoc sp. PCC 7120 | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, Apocytochrome f, ... | | Authors: | Craner, W.A, Baniulis, D, Yamashita, E. | | Deposit date: | 2008-09-27 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Function, Stability, and Chemical Modification of the Cyanobacterial Cytochrome b6f Complex from Nostoc sp. PCC 7120
J.Biol.Chem., 284, 2009
|
|
4UNU
 
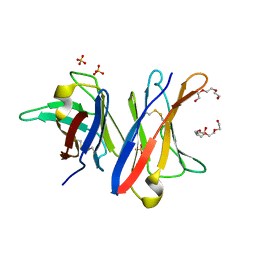 | | MCG - a dimer of lambda variable domains | | Descriptor: | IG LAMBDA CHAIN V-II REGION MGC, POLYETHYLENE GLYCOL (N=34), SULFATE ION | | Authors: | Brumshtein, B, Esswein, S.R, Landau, M, Ryan, C, Whitelegge, J, Casio, D, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2014-05-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Formation of Amyloid Fibers by Monomeric Light-Chain Variable Domains.
J.Biol.Chem., 289, 2014
|
|
4UNV
 
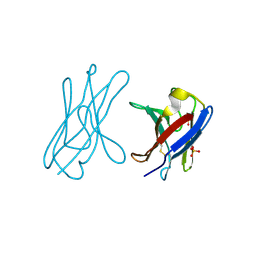 | | Covalent dimer of lambda variable domains | | Descriptor: | IG LAMBDA CHAIN V-II REGION MGC, SULFATE ION | | Authors: | Brumshtein, B, Esswein, S, Landau, M, Ryan, C, Whitelegge, J, Sawaya, M, Eisenberg, D.S. | | Deposit date: | 2014-05-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Formation of Amyloid Fibers by Monomeric Light-Chain Variable Domains.
J.Biol.Chem., 289, 2014
|
|
8ENY
 
 | |
8ENS
 
 | |
8ENX
 
 | |
8ENZ
 
 | |
8ENW
 
 | |
8EO0
 
 | |
4OLO
 
 | | Ligand-free structure of the GrpU microcompartment shell protein from Clostridiales bacterium 1_7_47FAA | | Descriptor: | BMC domain protein | | Authors: | Thompson, M.C, Ahmed, H, McCarty, K.N, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a unique fe-s cluster binding site in a glycyl-radical type microcompartment shell protein.
J.Mol.Biol., 426, 2014
|
|
4OLP
 
 | | Ligand-free structure of the GrpU microcompartment shell protein from Pectobacterium wasabiae | | Descriptor: | GrpU microcompartment shell protein | | Authors: | Wheatley, N.M, Thompson, M.C, Gidaniyan, S.D, Sawaya, M.R, Jorda, J, Yeates, T.O. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Identification of a unique fe-s cluster binding site in a glycyl-radical type microcompartment shell protein.
J.Mol.Biol., 426, 2014
|
|
4Y0L
 
 | | Mycobacterial membrane protein MmpL11D2 | | Descriptor: | IODIDE ION, Putative membrane protein mmpL11, SULFATE ION | | Authors: | Torres, R, Chim, N, Goulding, C.W. | | Deposit date: | 2015-02-06 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure and Interactions of Periplasmic Domains of Crucial MmpL Membrane Proteins from Mycobacterium tuberculosis.
Chem.Biol., 22, 2015
|
|
3COC
 
 | |
3COD
 
 | | Crystal Structure of T90A/D115A mutant of Bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Joh, N.H, Min, A, Faham, S, Bowie, J.U. | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins.
Nature, 453, 2008
|
|
3SGO
 
 | |
3SGS
 
 | |
