7AG0
 
 | | Complex between the bone morphogenetic protein 2 and its antagonist Noggin | | Descriptor: | Bone morphogenetic protein 2, GLYCEROL, Noggin | | Authors: | Robert, C, Bruck, F, Herman, R, Vandevenne, M, Filee, P, Kerff, F, Matagne, A. | | Deposit date: | 2020-09-21 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structural analysis of the interaction between human cytokine BMP-2 and the antagonist Noggin reveals molecular details of cell chondrogenesis inhibition.
J.Biol.Chem., 299, 2023
|
|
6SO0
 
 | | NMR solution structure of the family 14 carbohydrate binding module (CBM14) from human chitotriosidase | | Descriptor: | Chitotriosidase-1 | | Authors: | Madland, E, Crasson, O, Vandevenne, M, Sorlie, M, Aachmann, F.L. | | Deposit date: | 2019-08-28 | | Release date: | 2020-01-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR and Fluorescence Spectroscopies Reveal the Preorganized Binding Site in Family 14 Carbohydrate-Binding Module from Human Chitotriosidase.
Acs Omega, 4, 2019
|
|
2MF8
 
 | | HADDOCK model of MyT1 F4F5 - DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*AP*AP*AP*GP*TP*TP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*AP*AP*CP*TP*TP*TP*CP*GP*GP*T)-3'), Myelin transcription factor 1, ... | | Authors: | Gamsjaeger, R, O'Connell, M.R, Cubeddu, L, Shepherd, N.E, Lowry, J.A, Kwan, A.H, Vandevenne, M, Swanton, M.K, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A structural analysis of DNA binding by myelin transcription factor 1 double zinc fingers.
J.Biol.Chem., 288, 2013
|
|
4IS1
 
 | | Crystal structure of ZNF217 bound to DNA | | Descriptor: | 5'-D(*AP*AP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', 5'-D(*TP*TP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', CHLORIDE ION, ... | | Authors: | Vandevenne, M.S, Jacques, D.A, Guss, J.M, Mackay, J.P. | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into DNA recognition by zinc fingers revealed by structural analysis of the oncoprotein ZNF217.
J.Biol.Chem., 288, 2013
|
|
4ZV6
 
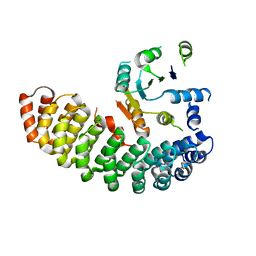 | | Crystal structure of the artificial alpharep-7 octarellinV.1 complex | | Descriptor: | AlphaRep-7, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Urvoas, A, Valerio-Lepiniec, M, Minard, P, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-18 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
7PA5
 
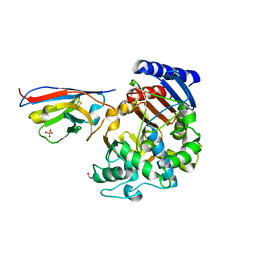 | | Complex between the beta-lactamase CMY-2 with an inhibitory nanobody | | Descriptor: | Beta-lactamase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Frederic Cawez, F.C, Frederic Kerff, F.K, Moreno Galleni, M.G, Raphael Herman, R.H. | | Deposit date: | 2021-07-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.184 Å) | | Cite: | Development of Nanobodies as Theranostic Agents against CMY-2-Like Class C beta-Lactamases.
Antimicrob.Agents Chemother., 67, 2023
|
|
5BOP
 
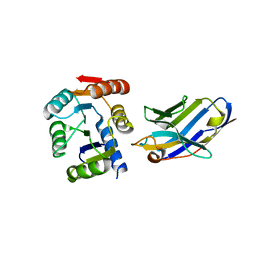 | | Crystal structure of the artificial nanobody octarellinV.1 complex | | Descriptor: | Nanobody, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Pardon, E, Steyaert, J, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-27 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
2N5N
 
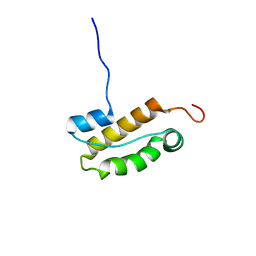 | | Structure of an N-terminal domain of CHD4 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4 | | Authors: | Silva, A.P.G, Mackay, J.P. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The N-terminal Region of Chromodomain Helicase DNA-binding Protein 4 (CHD4) Is Essential for Activity and Contains a High Mobility Group (HMG) Box-like-domain That Can Bind Poly(ADP-ribose).
J.Biol.Chem., 291, 2016
|
|
