1BQT
 
 | | THREE-DIMENSIONAL STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR-I (IGF-I) DETERMINED BY 1H-NMR AND DISTANCE GEOMETRY, 6 STRUCTURES | | Descriptor: | INSULIN-LIKE GROWTH FACTOR-I | | Authors: | Sato, A, Nishimura, S, Ohkubo, T, Kyogoku, Y, Koyama, S, Kobayashi, M, Yasuda, T, Kobayashi, Y. | | Deposit date: | 1998-08-18 | | Release date: | 1999-05-18 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of human insulin-like growth factor-I (IGF-I) determined by 1H-NMR and distance geometry.
Int.J.Pept.Protein Res., 41, 1993
|
|
2RRD
 
 | | Structure of HRDC domain from human Bloom syndrome protein, BLM | | Descriptor: | HRDC domain from Bloom syndrome protein | | Authors: | Sato, A, Mishima, M, Nagai, A, Kim, S.Y, Ito, Y, Hakoshima, T, Jee, J.G, Kitano, K. | | Deposit date: | 2010-07-19 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HRDC domain of human Bloom syndrome protein BLM
J.Biochem., 148, 2010
|
|
2RQL
 
 | |
1ID7
 
 | | SOLUTION STRUCTURE OF SYR6 | | Descriptor: | SYR6 | | Authors: | Sato, A, Kawaguchi, K, Kimura, K, Tanimura, R, Sone, S. | | Deposit date: | 2001-04-04 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A peptide mimetic of IFN, the first proof of a small peptidic agonist for heterodimeric cytokine receptor
To be Published
|
|
1ID6
 
 | | SOLUTION STRUCTURES OF SYR6 | | Descriptor: | SYR6 | | Authors: | Sato, A, Kawaguchi, K, Kimura, K, Tanimura, R, Sone, S. | | Deposit date: | 2001-04-04 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A peptide mimetic of IFN, the first proof of a small peptidic agonist for heterodimeric cytokine receptor
To be Published
|
|
1X5T
 
 | | Solution structure of the second RRM domain in splicing factor = 3B | | Descriptor: | Splicing factor 3B subunit 4 | | Authors: | Sato, A, Kuwasako, K, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-16 | | Release date: | 2005-11-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RRM domain in splicing factor = 3B
To be Published
|
|
1X5S
 
 | | Solution structure of RRM domain in A18 hnRNP | | Descriptor: | Cold-inducible RNA-binding protein | | Authors: | Sato, A, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-16 | | Release date: | 2005-11-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in A18 hnRNP
To be Published
|
|
1JMX
 
 | |
1JMZ
 
 | | crystal structure of a quinohemoprotein amine dehydrogenase from pseudomonas putida with inhibitor | | Descriptor: | Amine Dehydrogenase, HEME C, NICKEL (II) ION, ... | | Authors: | Satoh, A, Miyahara, I, Hirotsu, K. | | Deposit date: | 2001-07-20 | | Release date: | 2002-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of quinohemoprotein amine dehydrogenase from Pseudomonas putida. Identification of a novel quinone cofactor encaged by multiple thioether cross-bridges.
J.Biol.Chem., 277, 2002
|
|
1UDY
 
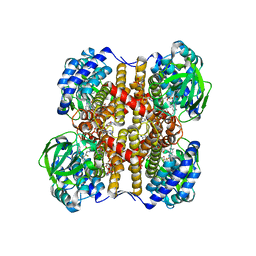 | | Medium-Chain Acyl-CoA Dehydrogenase with 3-Thiaoctanoyl-CoA | | Descriptor: | 3-THIAOCTANOYL-COENZYME A, Acyl-CoA dehydrogenase, medium-chain specific, ... | | Authors: | Satoh, A, Nakajima, Y, Miyahara, I, Hirotsu, K, Tanaka, T, Nishina, Y, Shiga, K, Tamaoki, H, Setoyama, C, Miura, R. | | Deposit date: | 2003-05-07 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the transition state analog of medium-chain acyl-CoA dehydrogenase. Crystallographic and molecular orbital studies on the charge-transfer complex of medium-chain acyl-CoA dehydrogenase with 3-thiaoctanoyl-CoA
J.BIOCHEM.(TOKYO), 134, 2003
|
|
3AG6
 
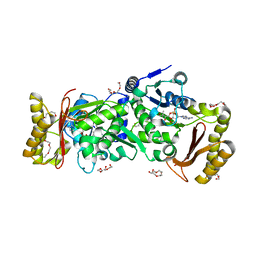 | | Crystal Structure of Pantothenate Synthetase from Staphylococcus aureus in complex with pantoyl adenylate | | Descriptor: | ACETIC ACID, PANTOYL ADENYLATE, Pantothenate synthetase, ... | | Authors: | Satoh, A, Konishi, S, Tamura, H, Stickland, H.G, Whitney, H.M, Smith, A.G, Matsumura, H, Inoue, T. | | Deposit date: | 2010-03-19 | | Release date: | 2010-07-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate-induced closing of the active site revealed by the crystal structure of pantothenate synthetase from Staphylococcus aureus.
Biochemistry, 49, 2010
|
|
3AG5
 
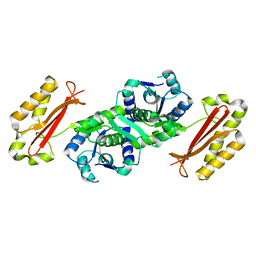 | | Crystal Structure of Pantothenate Synthetase from Staphylococcus aureus | | Descriptor: | Pantothenate synthetase | | Authors: | Satoh, A, Konishi, S, Tamura, H, Stickland, H.G, Whitney, H.M, Smith, A.G, Matsumura, H, Inoue, T. | | Deposit date: | 2010-03-19 | | Release date: | 2010-07-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate-induced closing of the active site revealed by the crystal structure of pantothenate synthetase from Staphylococcus aureus.
Biochemistry, 49, 2010
|
|
1QR3
 
 | | Structure of porcine pancreatic elastase in complex with FR901277, a novel macrocyclic inhibitor of elastases at 1.6 angstrom resolution | | Descriptor: | CALCIUM ION, Chymotrypsin-like elastase family member 1, FR901277 Inhibitor, ... | | Authors: | Nakanishi, I, Kinoshita, T, Sato, A, Tada, T. | | Deposit date: | 1999-06-18 | | Release date: | 2000-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of porcine pancreatic elastase complexed with FR901277, a novel macrocyclic inhibitor of elastases, at 1.6 A resolution.
Biopolymers, 53, 2000
|
|
8WO8
 
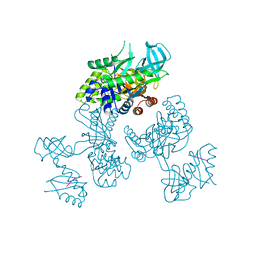 | | Crystal Structure of an RNA-binding protein, FAU-1, from Pyrococcus furiosus | | Descriptor: | Probable ribonuclease FAU-1, RNA (5'-R(P*AP*UP*A)-3') | | Authors: | Kawai, G, Okada, K, Baba, S, Sato, A, Sakamoto, T, Kanai, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Homo-trimeric structure of the ribonuclease for rRNA processing, FAU-1, from Pyrococcus furiosus.
J.Biochem., 175, 2024
|
|
5GVQ
 
 | | Solution structure of the first RRM domain of human spliceosomal protein SF3b49 | | Descriptor: | Splicing factor 3B subunit 4 | | Authors: | Kuwasako, K, Nameki, N, Tsuda, K, Takahashi, M, Sato, A, Tochio, N, Inoue, M, Terada, T, Kigawa, T, Kobayashi, N, Shirouzu, M, Ito, T, Sakamoto, T, Wakamatsu, K, Guntert, P, Takahashi, S, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first RNA recognition motif domain of human spliceosomal protein SF3b49 and its mode of interaction with a SF3b145 fragment.
Protein Sci., 26, 2017
|
|
3AJI
 
 | | Structure of Gankyrin-S6ATPase photo-cross-linked site-specifically, and incoporated by genetic code expansion | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, Proteasome (Prosome, macropain) 26S subunit, ... | | Authors: | Sato, S, Mimasu, S, Sato, A, Hino, N, Sakamoto, K, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-06-07 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic study of a site-specifically cross-linked protein complex with a genetically incorporated photoreactive amino acid
Biochemistry, 50, 2011
|
|
3ABG
 
 | | X-ray Crystal Analysis of Bilirubin Oxidase from Myrothecium verrucaria at 2.3 angstrom Resolution using a Twin Crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bilirubin oxidase, COPPER (II) ION, ... | | Authors: | Mizutani, K, Toyoda, M, Sagara, K, Takahashi, N, Sato, A, Kamitaka, Y, Tsujimura, S, Nakanishi, Y, Sugiura, T, Yamaguchi, S, Kano, K, Mikami, B. | | Deposit date: | 2009-12-10 | | Release date: | 2010-08-18 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray analysis of bilirubin oxidase from Myrothecium verrucaria at 2.3 A resolution using a twinned crystal
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1QPU
 
 | | SOLUTION STRUCTURE OF OXIDIZED ESCHERICHIA COLI CYTOCHROME B562 | | Descriptor: | CYTOCHROME B562, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Faraone-Mennella, J, Rosato, A, Barker, P.D, Fersht, A.R. | | Deposit date: | 1999-05-30 | | Release date: | 1999-06-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of oxidized Escherichia coli cytochrome b562.
Biochemistry, 38, 1999
|
|
2KMX
 
 | | Solution structure of the nucleotide binding domain of the human Menkes protein in the ATP-bound form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Inagaki, S, Migliardi, M, Rosato, A. | | Deposit date: | 2009-08-05 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The binding mode of ATP revealed by the solution structure of the N-domain of human ATP7A.
J.Biol.Chem., 2009
|
|
2KMV
 
 | | Solution structure of the nucleotide binding domain of the human Menkes protein in the ATP-free form | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Inagaki, S, Migliardi, M, Rosato, A. | | Deposit date: | 2009-08-05 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The binding mode of ATP revealed by the solution structure of the N-domain of human ATP7A.
J.Biol.Chem., 2009
|
|
2KIJ
 
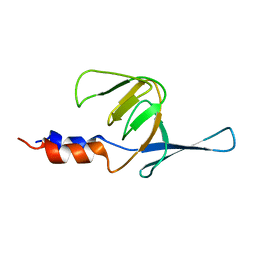 | | Solution structure of the Actuator domain of the copper-transporting ATPase ATP7A | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Nushi, F, Natile, G, Rosato, A. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the actuator domain of ATP7A and ATP7B, the Menkes and Wilson disease proteins
Biochemistry, 48, 2009
|
|
1BD6
 
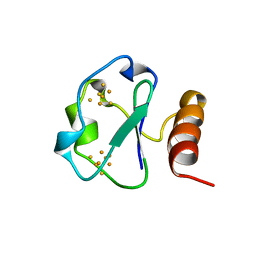 | | 7-FE FERREDOXIN FROM BACILLUS SCHLEGELII, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | 7-FE FERREDOXIN, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Aono, S, Bentrop, D, Bertini, I, Donaire, A, Luchinat, C, Niikura, Y, Rosato, A. | | Deposit date: | 1998-05-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized Fe7S8 ferredoxin from the thermophilic bacterium Bacillus schlegelii by 1H NMR spectroscopy.
Biochemistry, 37, 1998
|
|
1BC6
 
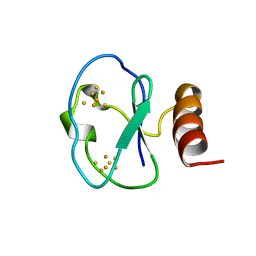 | | 7-FE FERREDOXIN FROM BACILLUS SCHLEGELII, NMR, 20 STRUCTURES | | Descriptor: | 7-FE FERREDOXIN, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Aono, S, Bentrop, D, Bertini, I, Donaire, A, Luchinat, C, Niikura, Y, Rosato, A. | | Deposit date: | 1998-05-05 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized Fe7S8 ferredoxin from the thermophilic bacterium Bacillus schlegelii by 1H NMR spectroscopy.
Biochemistry, 37, 1998
|
|
1N9C
 
 | |
3CJK
 
 | | Crystal structure of the adduct HAH1-Cd(II)-MNK1. | | Descriptor: | CADMIUM ION, Copper transport protein ATOX1, Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Calderone, V, Felli, I, Della-Malva, N, Pavelkova, A, Rosato, A. | | Deposit date: | 2008-03-13 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Copper(I)-mediated protein-protein interactions result from suboptimal interaction surfaces.
Biochem.J., 422, 2009
|
|
