5X9J
 
 | |
5GK2
 
 | |
5GK0
 
 | |
5GK1
 
 | | Crystal structure of the ketosynthase StlD complexed with substrate | | Descriptor: | 3-OXO-5-METHYLHEXANOIC ACID, Ketosynthase StlD | | Authors: | Mori, T, Saito, Y, Morita, H, Abe, I. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Insight into the Enzymatic Formation of Bacterial Stilbene.
Cell Chem Biol, 23, 2016
|
|
5WQF
 
 | | Structure of fungal meroterpenoid isomerase Trt14 | | Descriptor: | CALCIUM ION, Isomerase trt14, N-PROPANOL | | Authors: | Mori, T, Iwabuchi, T, Matsuda, Y, Abe, I. | | Deposit date: | 2016-11-26 | | Release date: | 2017-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Molecular basis for the unusual ring reconstruction in fungal meroterpenoid biogenesis
Nat. Chem. Biol., 13, 2017
|
|
5WQH
 
 | | Structure of fungal meroterpenoid isomerase Trt14 complexed with substrate analog and endo-terretonin D | | Descriptor: | CALCIUM ION, Isomerase trt14, methyl (2S,4aR,4bS,5S,6aS,10aS,10bS,12aS)-2,4b,7,7,10a,12,12a-heptamethyl-5-oxidanyl-1,4,6,8-tetrakis(oxidanylidene)-4a,5,6a,9,10,10b-hexahydronaphtho[1,2-h]isochromene-2-carboxylate, ... | | Authors: | Mori, T, Matsuda, Y, Abe, I. | | Deposit date: | 2016-11-26 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Molecular basis for the unusual ring reconstruction in fungal meroterpenoid biogenesis
Nat. Chem. Biol., 13, 2017
|
|
5WQG
 
 | | Structure of fungal meroterpenoid isomerase Trt14 complexed with terretonin D | | Descriptor: | CALCIUM ION, Isomerase trt14, methyl (2S,4aR,4bS,5S,6aS,10aS,10bS,12aS)-2,4b,7,7,10a,12a-hexamethyl-12-methylidene-5-oxidanyl-1,4,6,8-tetrakis(oxidanylidene) -5,6a,9,10,10b,11-hexahydro-4aH-naphtho[1,2-h]isochromene-2-carboxylate | | Authors: | Mori, T, Matsuda, Y, Abe, I. | | Deposit date: | 2016-11-26 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the unusual ring reconstruction in fungal meroterpenoid biogenesis
Nat. Chem. Biol., 13, 2017
|
|
5WQI
 
 | | Structure of fungal meroterpenoid isomerase Trt14 complexed with hydrolyzed product | | Descriptor: | (1R,2S,4aS,4bS,8aS,10S,10aS)-2-[(2S)-3-methoxy-2-methyl-2-oxidanyl-3-oxidanylidene-propanoyl]-2,3,4b,8,8,10a-hexamethyl-10-oxidanyl-7,9-bis(oxidanylidene)-1,4a,5,6,8a,10-hexahydrophenanthrene-1-carboxylic acid, CALCIUM ION, Isomerase trt14 | | Authors: | Mori, T, Matsuda, Y, Abe, I. | | Deposit date: | 2016-11-27 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Molecular basis for the unusual ring reconstruction in fungal meroterpenoid biogenesis
Nat. Chem. Biol., 13, 2017
|
|
8Y9P
 
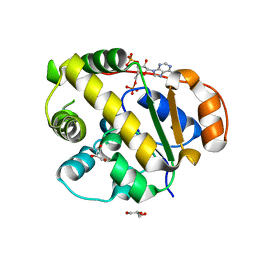 | |
7EXZ
 
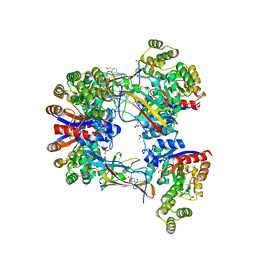 | | DgpB-DgpC complex apo 2.5 angstrom | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AP_endonuc_2 domain-containing protein, DgpB, ... | | Authors: | Mori, T, Senda, M, Senda, T, Abe, I. | | Deposit date: | 2021-05-29 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7EXB
 
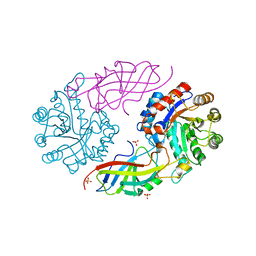 | | DfgA-DfgB complex apo 2.4 angstrom | | Descriptor: | DfgB, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Mori, T, Senda, M, Senda, T, Abe, I. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7DRE
 
 | | Cryo-EM structure of DfgA-B at 2.54 angstrom resolution | | Descriptor: | DfgB, Sugar phosphate isomerase/epimerase | | Authors: | Mori, T, Moriya, T, Adachi, N, Senda, T, Abe, I. | | Deposit date: | 2020-12-28 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7DRD
 
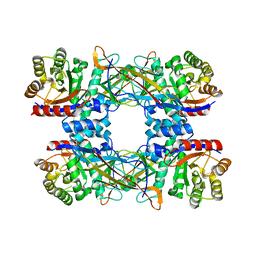 | | Cryo-EM structure of DgpB-C at 2.85 angstrom resolution | | Descriptor: | AP_endonuc_2 domain-containing protein, DgpB | | Authors: | Mori, T, Moriya, T, Adachi, N, Senda, T, Abe, I. | | Deposit date: | 2020-12-28 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
8K3H
 
 | |
8K3I
 
 | |
7BVR
 
 | | DgpB-DgpC complex apo | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AP_endonuc_2 domain-containing protein, DgpB, ... | | Authors: | Mori, T, He, H, Abe, I. | | Deposit date: | 2020-04-11 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7BVS
 
 | | DfgA-DfgB complex apo | | Descriptor: | DfgB, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Mori, T, He, H, Abe, I. | | Deposit date: | 2020-04-11 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7YN3
 
 | | Crystal structure of CcbD complex with CcbZ carrier protein domain | | Descriptor: | 1,1'-ethane-1,2-diyldipyrrolidine-2,5-dione, 4'-PHOSPHOPANTETHEINE, CcbD, ... | | Authors: | Mori, T, Lyu, S, Abe, I. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of CcbD complex with carrier protein
To Be Published
|
|
7YN1
 
 | |
7YN2
 
 | | Crystal structure of CcbD with methylthiolincosamide | | Descriptor: | (2R,3R,4S,5R,6R)-2-[(1R,2R)-1-azanyl-2-oxidanyl-propyl]-6-methylsulfanyl-oxane-3,4,5-triol, CcbD | | Authors: | Mori, T, Lyu, S, Abe, I. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CcbD with methylthiolincosamide
To Be Published
|
|
4YZJ
 
 | |
4YLA
 
 | | Crystal structure of the indole prenyltransferase MpnD complexed with indolactam V and DMSPP | | Descriptor: | (2S,5S)-5-(hydroxymethyl)-1-methyl-2-(propan-2-yl)-1,2,4,5,6,8-hexahydro-3H-[1,4]diazonino[7,6,5-cd]indol-3-one, Aromatic prenyltransferase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Mori, T, Morita, H, Abe, I. | | Deposit date: | 2015-03-05 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases.
Nat Commun, 7, 2016
|
|
4YZL
 
 | | Crystal structure of the indole prenyltransferase TleC complexed with indolactam V and DMSPP | | Descriptor: | (2S,5S)-5-(hydroxymethyl)-1-methyl-2-(propan-2-yl)-1,2,4,5,6,8-hexahydro-3H-[1,4]diazonino[7,6,5-cd]indol-3-one, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Tryptophan dimethylallyltransferase | | Authors: | Mori, T, Morita, H, Abe, I. | | Deposit date: | 2015-03-25 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases.
Nat Commun, 7, 2016
|
|
3X27
 
 | | Structure of McbB in complex with tryptophan | | Descriptor: | Cucumopine synthase, TRYPTOPHAN | | Authors: | Mori, T, Sahashi, S, Morita, H, Abe, I. | | Deposit date: | 2014-12-10 | | Release date: | 2015-10-28 | | Last modified: | 2015-11-04 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Structural Basis for beta-Carboline Alkaloid Production by the Microbial Homodimeric Enzyme McbB
Chem.Biol., 22, 2015
|
|
4YL7
 
 | |
