1E8J
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS ZINC RUBREDOXIN, NMR, 20 STRUCTURES | | Descriptor: | RUBREDOXIN | | Authors: | Lamosa, P, Brennan, L, Vis, H, Turner, D.L, Santos, H. | | Deposit date: | 2000-09-21 | | Release date: | 2001-10-18 | | Last modified: | 2019-10-09 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Desulfovibrio gigas rubredoxin: a model for studying protein stabilization by compatible solutes.
Extremophiles, 5, 2001
|
|
1SPW
 
 | | Solution Structure of a Loop Truncated Mutant from D. gigas Rubredoxin, NMR | | Descriptor: | Rubredoxin | | Authors: | Pais, T.M, Lamosa, P, dos Santos, W, LeGall, J, Turner, D.L, Santos, H. | | Deposit date: | 2004-03-17 | | Release date: | 2005-03-29 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of protein stabilization by solutes: the importance of the hairpin loop in rubredoxins
FEBS J., 272, 2005
|
|
2LGN
 
 | | Lactococcin 972 | | Descriptor: | Lactococcin 972 | | Authors: | Turner, D.L, Lamosa, P, Martinez, B. | | Deposit date: | 2011-07-29 | | Release date: | 2012-08-01 | | Method: | SOLUTION NMR | | Cite: | Structure and properties of lactococcin 972 from Lactococcus lactis
To be Published
|
|
2LKV
 
 | | Staphylococcal Nuclease PHS variant | | Descriptor: | Thermonuclease | | Authors: | Matzapetakis, M, Pais, T.M, Lamosa, P, Turner, D.L, Santos, H. | | Deposit date: | 2011-10-21 | | Release date: | 2012-09-12 | | Method: | SOLUTION NMR | | Cite: | Mannosylglycerate stabilizes staphylococcal nuclease with restriction of slow beta-sheet motions.
Protein Sci., 21, 2012
|
|
4P2V
 
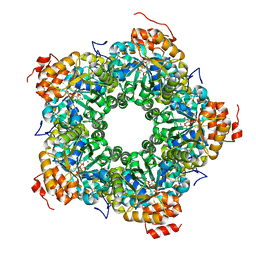 | | Structure of the AI-2 processing enzyme LsrF in complex with the product of the LsrG reaction P-HPD | | Descriptor: | (3R)-3-hydroxy-2,4-dioxopentyl dihydrogen phosphate, Uncharacterized aldolase LsrF | | Authors: | Miller, S.T, Oh, I.K, Xavier, K.B. | | Deposit date: | 2014-03-04 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | LsrF, a coenzyme A-dependent thiolase, catalyzes the terminal step in processing the quorum sensing signal autoinducer-2.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3QMQ
 
 | | Crystal Structure of E. coli LsrG | | Descriptor: | Autoinducer-2 (AI-2) modifying protein LsrG | | Authors: | Miller, S.T, Russell, C. | | Deposit date: | 2011-02-04 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Processing the Interspecies Quorum-sensing Signal Autoinducer-2 (AI-2): CHARACTERIZATION OF PHOSPHO-(S)-4,5-DIHYDROXY-2,3-PENTANEDIONE ISOMERIZATION BY LsrG PROTEIN.
J.Biol.Chem., 286, 2011
|
|
4D02
 
 | | The crystallographic structure of Flavorubredoxin from Escherichia coli | | Descriptor: | ANAEROBIC NITRIC OXIDE REDUCTASE FLAVORUBREDOXIN, CHLORIDE ION, FE (III) ION, ... | | Authors: | Romao, C.V, Vicente, J.B, Bandeiras, T, Carrondo, M.A, Teixeira, M, Frazao, C. | | Deposit date: | 2014-04-23 | | Release date: | 2014-09-03 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Structure of Escherichia Coli Flavodiiron Nitric Oxide Reductase.
J.Mol.Biol., 428, 2016
|
|
5LMC
 
 | | Oxidized flavodiiron core of Escherichia coli flavorubredoxin, including the Fe-4SG atoms from its rubredoxin domain | | Descriptor: | ACETIC ACID, Anaerobic nitric oxide reductase flavorubredoxin, CACODYLATE ION, ... | | Authors: | Romao, C.V, Borges, P.T, Vicente, J.B, Carrondo, M.A, Teixeira, M, Frazao, C. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Escherichia coli Flavodiiron Nitric Oxide Reductase.
J.Mol.Biol., 428, 2016
|
|
5LLD
 
 | | Flavodiiron core of Escherichia coli flavorubredoxin in the reduced form. | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Romao, C.V, Borges, P.T, Vicente, J.B, Carrondo, M.A, Teixeira, M, Frazao, C. | | Deposit date: | 2016-07-27 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structure of Escherichia coli Flavodiiron Nitric Oxide Reductase.
J.Mol.Biol., 428, 2016
|
|
