6JYI
 
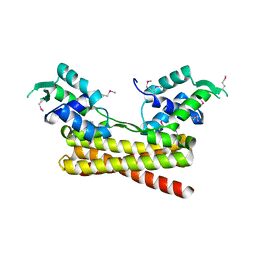 | | Crystal structure of the PadR-like transcriptional regulator BC1756 from Bacillus cereus | | Descriptor: | Transcriptional repressor PadR | | Authors: | Kim, T.H, Park, S.C, Lee, K.C, Song, W.S, Yoon, S.I. | | Deposit date: | 2019-04-26 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and DNA-binding studies of the PadR-like transcriptional regulator BC1756 from Bacillus cereus.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
5SWN
 
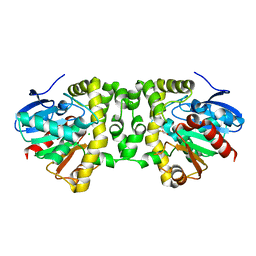 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Fluoroacetate - Cocrystallized | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5T4T
 
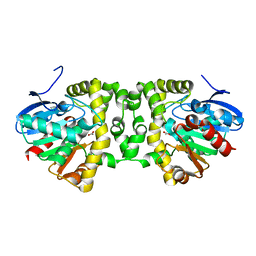 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn - Apo No Halide | | Descriptor: | ACETATE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-08-30 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3A
 
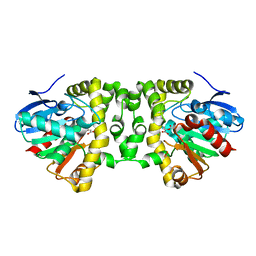 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/Fluoroacetate - Cocrystallized - Both Protomers Reacted with Ligand | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3B
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Chloroacetate - Cocrystallized | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, chloroacetic acid | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3D
 
 | |
5K3C
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - WT/5-Fluorotryptophan | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3F
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/Fluoroacetate - Cocrystallized - Single Protomer Reacted with Ligand | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3E
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Glycolate - Cocrystallized | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, GLYCOLIC ACID | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
6QKU
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Chloroacetate soaked 2hr | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, GLYCOLIC ACID, ... | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
6QKS
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Apo | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
6QKT
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Fluoroacetate soaked 24hr - Glycolate bound | | Descriptor: | Fluoroacetate dehalogenase, GLYCOLIC ACID | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.512 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
6QKW
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Fluoroacetate soaked 2hr | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, GLYCOLIC ACID, ... | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.512 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
5JQH
 
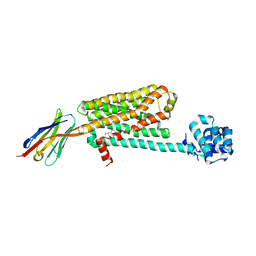 | | Structure of beta2 adrenoceptor bound to carazolol and inactive-state stabilizing nanobody, Nb60 | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, CHOLESTEROL, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Staus, D.P, Strachan, R.T, Manglik, A, Pani, B, Kahsai, A.W, Kim, T.H, Wingler, L.M, Ahn, S, Chatterjee, A, Masoudi, A, Kruse, A.C, Pardon, E, Steyaert, J, Weis, W.I, Prosser, R.S, Kobilka, B.K, Costa, T, Lefkowitz, R.J. | | Deposit date: | 2016-05-05 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation.
Nature, 535, 2016
|
|
7BYK
 
 | |
5ZIY
 
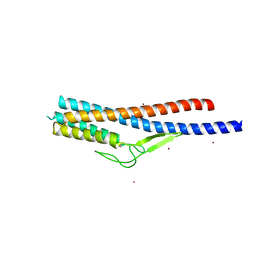 | | Crystal structure of Bacillus cereus FlgL | | Descriptor: | Flagellar hook-associated protein 3, ZINC ION | | Authors: | Hong, H.J, Kim, T.H, Song, W.S, Yoon, S.I. | | Deposit date: | 2018-03-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FlgL and its implications for flagellar assembly
Sci Rep, 8, 2018
|
|
5ZIZ
 
 | |
5ZJ0
 
 | |
8KA0
 
 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-bound calmodulin and a nicotinamide adenine dinucleotide (NAD+) | | Descriptor: | CALCIUM ION, Calmodulin-2, GLYCEROL, ... | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
8KA2
 
 | |
8K9Z
 
 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-bound calmodulin | | Descriptor: | CALCIUM ION, Calmodulin-2, RDTND-RID CBD | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
8KA1
 
 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-free calmodulin | | Descriptor: | Calmodulin-2, MAGNESIUM ION, RDTND-RID CBD | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
5ZCU
 
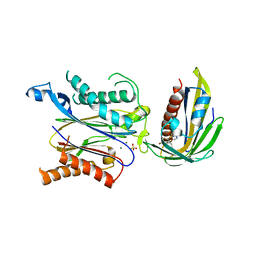 | | Crystal structure of RCAR3:PP2C wild-type with pyrabactin | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Han, S, Lee, Y, Lee, S. | | Deposit date: | 2018-02-20 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structural determinants for pyrabactin recognition in ABA receptors in Oryza sativa.
Plant Mol.Biol., 100, 2019
|
|
5ZCH
 
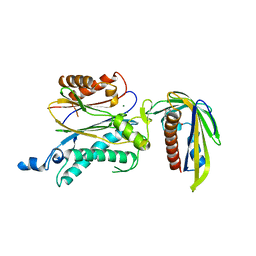 | | Crystal structure of OsPP2C50 I267W:OsPYL/RCAR3 with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION, ... | | Authors: | Lee, S, Han, S. | | Deposit date: | 2018-02-17 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.474 Å) | | Cite: | Comprehensive survey of the VxG Phi L motif of PP2Cs from Oryza sativa reveals the critical role of the fourth position in regulation of ABA responsiveness.
Plant Mol.Biol., 101, 2019
|
|
5ZCG
 
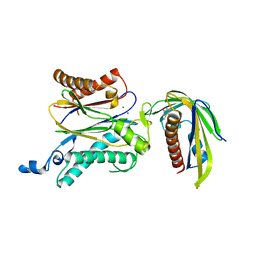 | | Crystal structure of OsPP2C50 S265L/I267V:OsPYL/RCAR3 with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Lee, S, Han, S. | | Deposit date: | 2018-02-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comprehensive survey of the VxG Phi L motif of PP2Cs from Oryza sativa reveals the critical role of the fourth position in regulation of ABA responsiveness.
Plant Mol.Biol., 101, 2019
|
|
